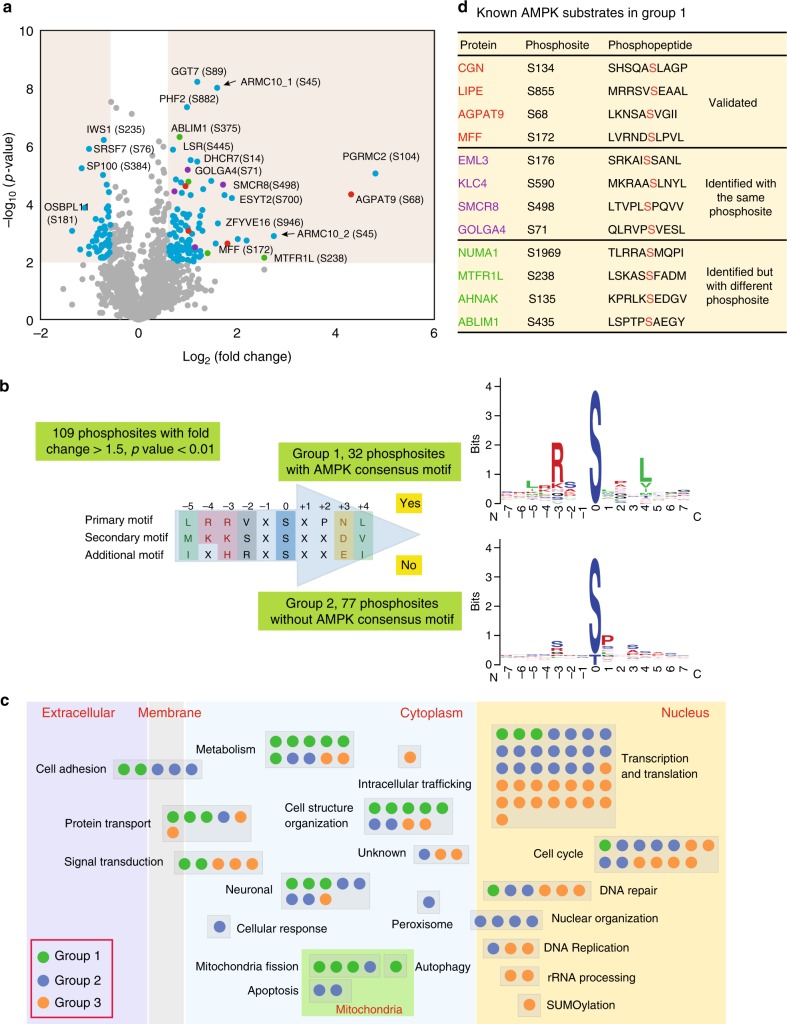
Fig. 2.
Bioinformatics analysis of the phosphoproteomic results. a Volcano plot of quantitative analysis of phosphopeptides identified by MS. Peptides with fold-changes (WT AMPK:AMPKα1/α2-DKO ratio) >1.5 (p < 0.01, t-test) were selected as peptides phosphorylated at markedly higher levels in WT cells than in AMPKα1/α2-DKO cells. Red dots are the AMPK phosphorylation substrates that had previously been validated; purple dots are peptides identified as AMPK phosphorylation substrates but not validated; green dots are peptides identified as AMPK phosphorylation substrates by previous study, but not the same phosphorylation sites; blue dots are other AMPK phosphorylation substrates identified by our experiments; gray dots are the phosphorylation peptides identified by this study but showing no significant difference between AMPKα1/α2-DKO and WT samples. The black arrows indicate the phosphopeptides from ARMC10, which we studied further. Please note that these two phosphopeptides are from the same S45 phosphorylation site in two different ARMC10 isoforms (i.e., ARMC10_1 is in a shorter isoform of 308 residues and ARMC10_2 is in a longer isoform of 343 residues). b Logo motif of the phosphosites. The 109 phosphosites phosphorylated at higher levels in WT AMPK than in AMPKα1/α2-DKO cells were separated into groups 1 (with the conserved AMPK substrate motif) and 2 (without the motif). The phosphosites in these two groups were further analyzed by using WebLogo to create the sequence logos. c Bioinformatic analysis of 160 AMPK-regulated phosphosites. The 109 phosphorylated proteins in groups 1 or 2, as well as the 51 in group 3 (phosphorylated at significantly lower levels in WT AMPK cells than in AMPKα1/α2-DKO cells) were analyzed individually for their potential functions with Ingenuity Pathway Analysis and reference mining. Each dot in the figure represents one protein. d Known AMPK substrates in group 1. These substrates comprised three categories: validated, identified with high-throughput methods but not validated and identified as a substrate but with a different phosphosite