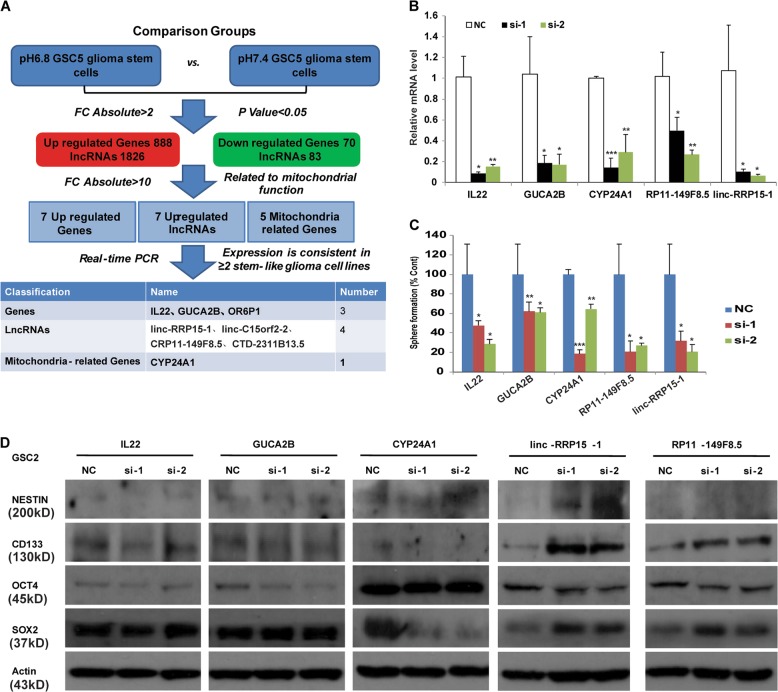
Fig. 2. Screening three genes and two lncRNAs that were changed under acidosis.
a Principle and process of screening. pH 7.4-treated and pH 6.8-treated GSC5 cells were used to conduct microarray analysis. Relative quantitative real-time PCR were used to verify the expression of mRNAs and lncRNAs. The selected eight genes and lncRNAs were shown (bottom panel). b Relative RNA levels of IL22, GUCA2B, CYP24A1, and lncRNA RP11-149F8.5, linc-RRP15-1 transfected with targeting siRNA in GSC2 cells (*P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). c Neurosphere formation ability of SLCs when knockdown of IL22, GUCA2B, CYP24A1, and lncRNA RP11-149 f8. 5, linc-RRP15-1 using siRNA. Left: Neurosphere formation assay showed the number of neurospheres (diameters larger than 50 µm) formed from GSC2 cells (*P < 0.05; **P < 0.01; ***P < 0.001, Student’s t-test). Right: phase contrast photomicrographs showed morphological change of GSC2 cells after knockdown of GUCA2B (bar = 100 μm, left; bar = 50 μm, right). d Immunoblotting of the expression of stemness markers NESTIN, CD133, OCT4, and SOX2 in U251-SLC that transfected with targeting siRNA of IL22, GUCA2B, CYP24A1, and lncRNA RP11-149 f8. 5, linc-RRP15-1