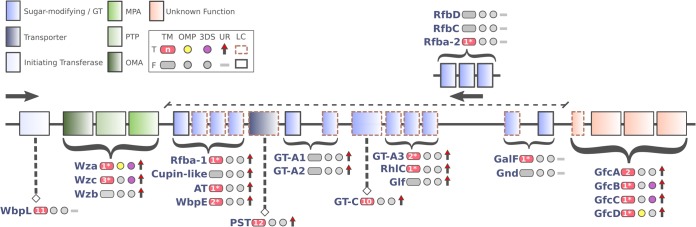
FIG 5.
Schematic organization of GFC region organization in Pcb1692. At the top left, the color assignments of known or unknown protein functionalities are shown using the following abbreviations: MPA, membrane-periplasmic auxiliary protein; PTP, protein tyrosine phosphatase; OMA, outer membrane auxiliary protein; GT, glycosyltransferase. A second box, framed by continuous lines, shows a binary table with graphical representations for the presence (T, true) or absence (F, false) of five parameters analyzed in the protein sequences. Abbreviations: TM, transmembrane segments; OMP, outer membrane protein; 3DS, available three-dimensional structure; UR, significant infection-induced upregulation in Pcb1692; LC, low conservation among SRE (detailed below), as assessed by synteny and sequence similarity. The number of detected TMs is within the pink shape. Numbers with an asterisk indicate the prediction methods diverge, whereas the lack of an asterisk indicates these methods converge in the number of TMs predicted. LC is attributed to sequences exhibiting fewer than 40 and 70 positive hits, respectively, in protein similarity (BLASTP) and synteny-based (MCScanX) pairwise comparisons using 100 SRE strains (see Table S6.1 for details). The arrows indicate the transcriptional orientation (sense or antisense) of the genes within the region, and the curly braces indicate ranges of genes transcribed in a given orientation for which gene names are displayed (top to bottom) according to the genomic order. A dashed line above the region representation highlights the serotype-specific block found between wza-wzb-wzc and gfcABCD in Pcb1692.