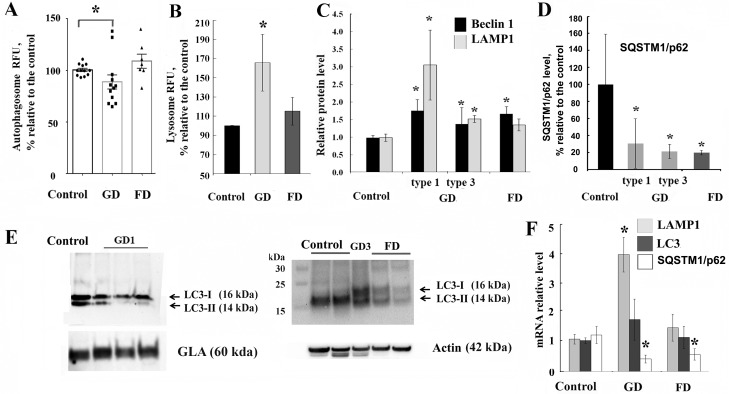
Fig 1. Autophagy-lysosomal complex is affected in GD and FD.
(A) PBMCs from healthy subjects (control), patients with GD (types 1 and 3) and FD were stained with Cyto-ID autophagy detection kit. The graph shows the relative levels of autophagosome vesicle number in control, GD and FD groups. Values were normalized to the control group. Each dot represents a patient sample.*p<0.05 One-way ANOVA, p = 0.016 ANOVA with Bartlett’s multiple comparisons test. (B) GD, FD and control PBMCs were stained with LysoTrackerRed to measure lysosomal accumulation. Values were normalized to the control group. GD (n = 5), FD (n = 6) and control (n = 10). *p<0.05. (C) Quantification of Beclin 1 and LAMP1 protein level from control (n = 10), GD1 (n = 5), GD3 (n = 4) and FD (n = 8) samples after western blot. The protein level was measured relative to total protein level. Values were normalized to the control group. *p<0.05 Student’s T-Test. (D) SQSTM1/p62 level in GD type 1 and type 3 (n = 11), FD (n = 5) and control (n = 5) samples were measured with a SQSTM1/p62 ELISA kit. SQSTM1/p62 level was measured in triplicates, on three biological repeats. Values normalized to control group. *p<0.05 Student’s T-Test. (E) Representative western blot showing LC3-I and LC3-II levels in PBMCs from GD (Type 1 and 3), FD and control samples. (F) Q-PCR was performed to determine relative LAMP1, LC3A/B and SQSTM1/p62 mRNA expression levels in GD and FD PBMCs. RNA level was measured in triplicates, n = 5 or 10 samples analysed each group. All data were expressed as S.E.M. and * p<0.05 vs control.