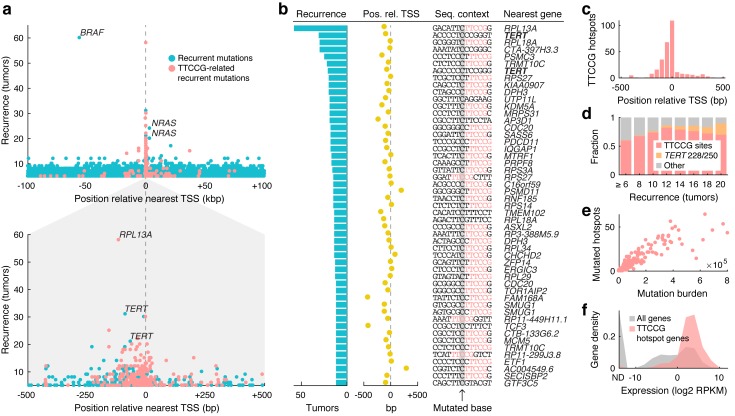
Fig 1. Widespread contribution from TTCCG-related sites to recurrent non-coding mutations in 221 whole melanoma genomes.
(a) Highly recurrent somatic mutations (individual genomic bases) aggregate near annotated transcription start sites (TSS) and typically colocalize with TTCCG elements. Recurrent sites having a TTCCG element within a +/-10 bp context on the mutated (pyrimidine) strand are indicated (red). The distance to the nearest TSS (x-axis) is adjusted for transcriptional orientation (upstream positioning of BRAF V600E mutations is explained by their relative proximity to the neighboring NDUFB2 promoter). Bottom panel: +/-500 bp close-up around the TSS. (b) Top 51 recurrent promoter sites (+/-500 bp), all mutated in ≥12/221 tumors (>5%). Degree of recurrence, position relative to TSS, sequence context with TTCCG highlighted in red, and nearest gene are indicated. (c) Positional distribution of TTCCG-related mutation hotspots near TSSs, based on 291 promoter sites recurrent in ≥5 tumors. (d) Proportion of recurrent promoter mutations (+/- 500 bp) that are TTCCG-related (red), TERT activating mutations (C228T/C250T; orange) or other (gray), as a function of recurrence. (e) Number of mutated TTCCG promoter hotspot sites per tumor, out of 291 in total as defined above, plotted against the whole-genome mutational burden across 221 melanomas. (f) TTCCG-related promoter hotspots arise preferably near highly expressed genes. 241 genes hosting 291 sites as defined above were considered. Expression levels were based on the median RPKM value across a subset of 38 TCGA melanomas with available RNA-seq. ND, not detected.