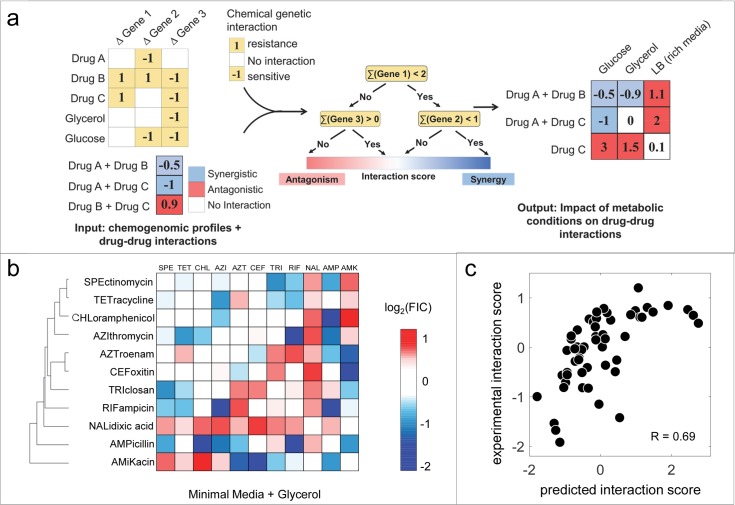
Fig 3. Predicting impact of novel metabolic environments on drug interactions using MAGENTA.
(a) Schematic workflow of MAGENTA approach. MAGENTA takes drug chemogenomic profiles and interactions among drugs or between drugs and metabolic conditions as input. This is used to train a Random Forest model which can predict synergy and antagonism among pair-wise or higher-order combinations of new drugs in novel metabolic conditions given their chemogenomic profiles. (b) All pairwise interactions among 11 antibiotics in minimal media supplemented with glycerol are represented as a heat map. Blue, white or red boxes correspond to synergistic, additive or antagonistic combinations. Clustering of interaction scores was done using Euclidean distance and average linkage (Unweighted average distance (UPGMA)). (c) Scatter plot comparison of MAGENTA predictions and experimental measurements in glycerol minimal media demonstrate that MAGENTA can robustly predict antibiotic synergy and antagonism in new environments (rank correlation R = 0.69, p = 1x10-8).