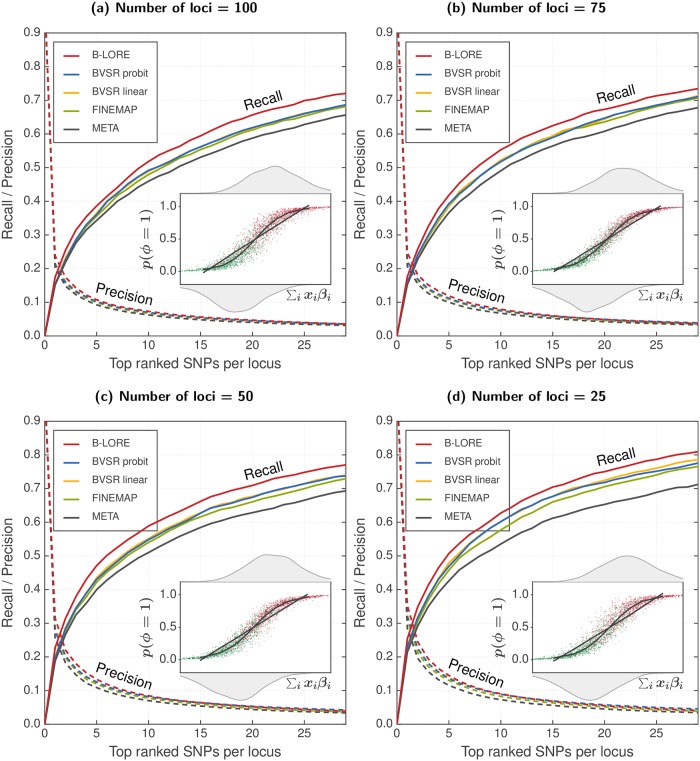
Fig 3. The advantage of B-LORE does not depend on the number of loci used for estimation.
Panels (a)—(d) show results from simulations using 25, 50, 75 and 100 loci respectively. As described in the main text, we used 13082 samples and each locus had ∼200 SNPs. All simulations used total heritability of and hence the heritability per locus is different for the different panels. We compared the ranking of SNPs at each locus using recall (solid lines, left y-axis) and precision (dotted lines, right y-axis), which were averaged over the loci and the simulation replicates. All methods were run with a maximum of two causal SNPs per locus. Different panels show the results at different number of loci. Insets schematically compare logistic model with linear model in one simulation (see Fig 1 for details). The heritability per locus increases when the number of loci is reduced. Multiple regression becomes increasingly better than single SNP analysis, but the advantage of B-LORE over other multiple regression methods does not change with the number of loci. Note also that the comparison between logistic model and the linear model in the insets does not change with the number of loci.