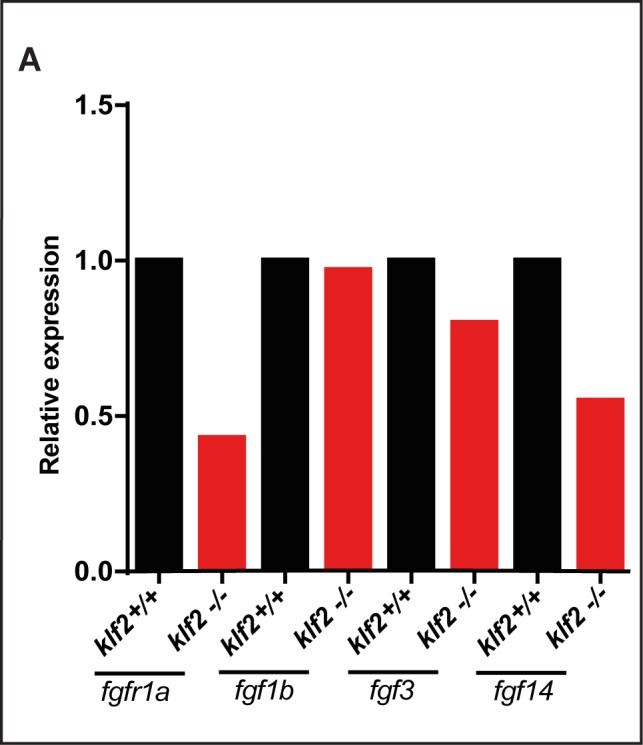
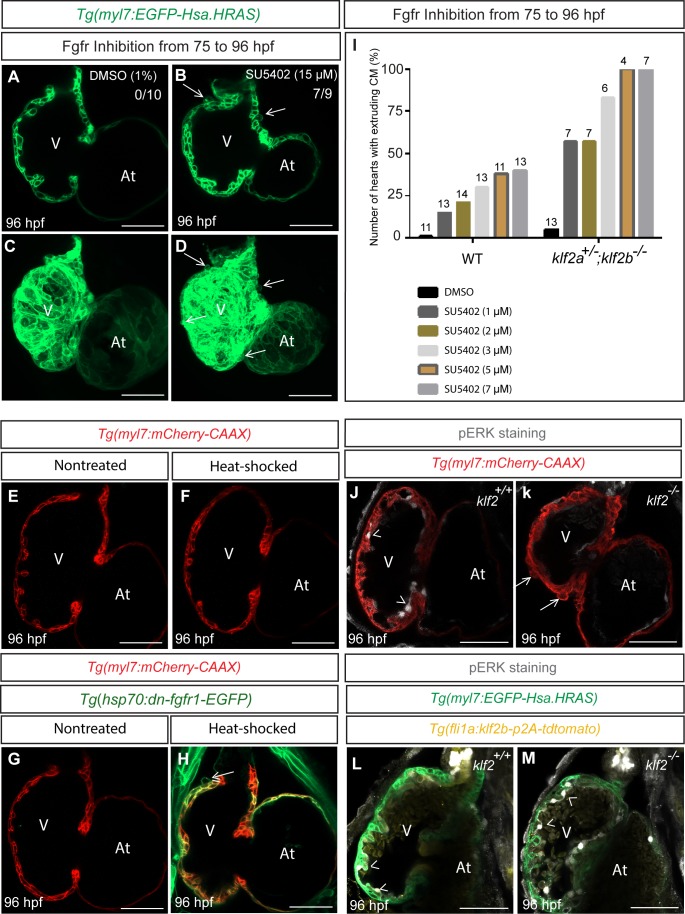
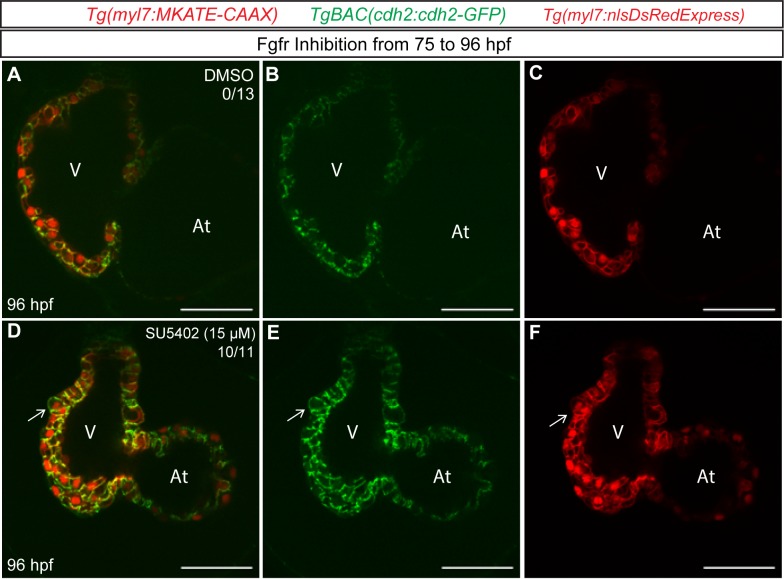
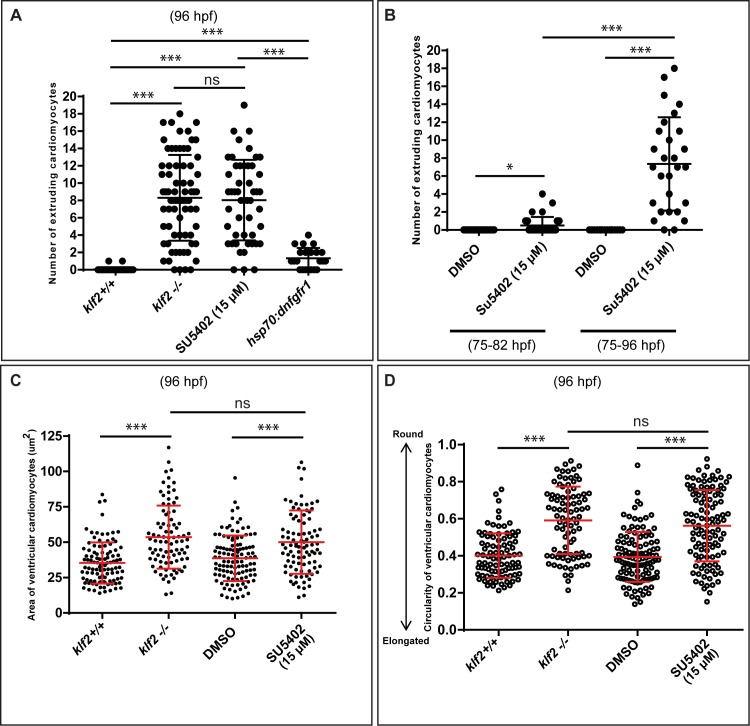
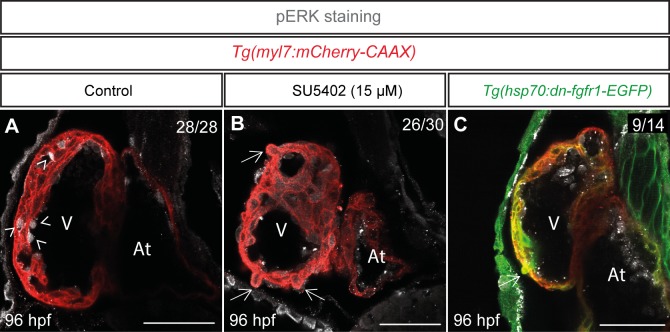
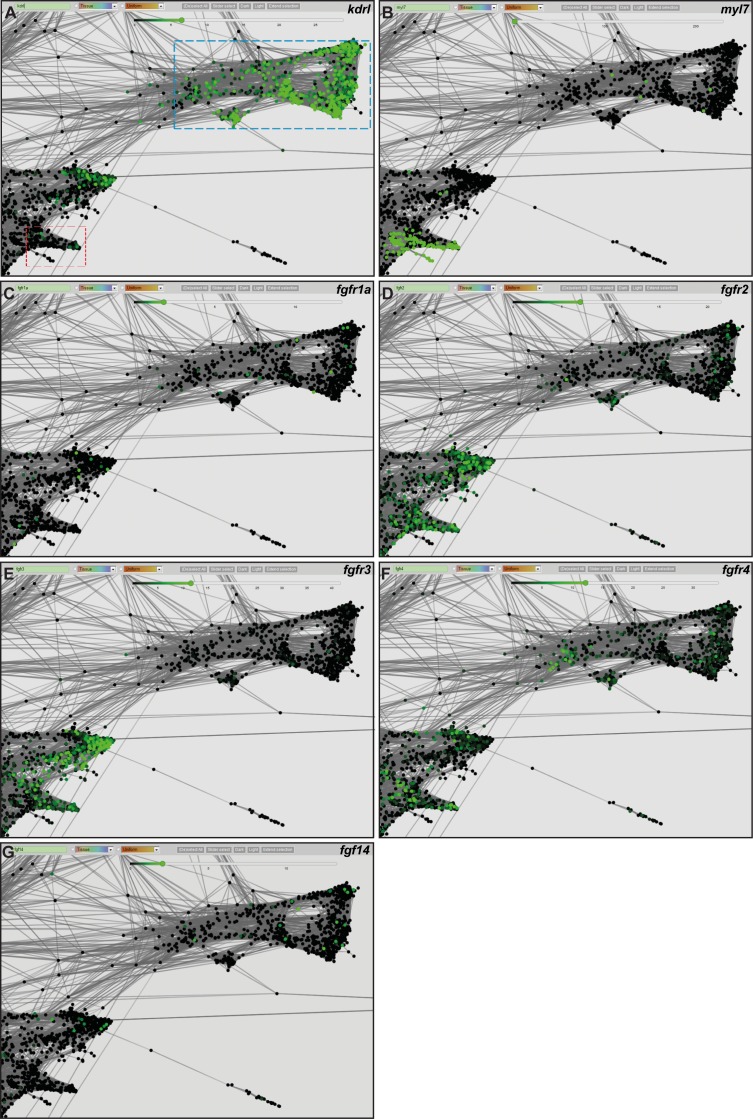
Figure 6. Inhibition of Fgfr signaling can lead to cardiomyocyte extrusion in WT animals.
(A–D) Confocal images of 96 hpf hearts; WT animals treated with DMSO as a control or FGFR inhibitor (SU5402) from 75 to 96 hpf; maximum intensity projections of hearts in (A) and (D) are shown in (C) and (D) respectively; arrows point to extruding cardiomyocytes. (E–H) 75 hpf Tg(myl7: mCherry-CAAX) (E–F) or Tg(hsp70:dn-fgfr1-EGFP);Tg(myl7: mCherry-CAAX) (G–H) animals were heat-stressed at 39°C for 1 hr (F and H) and their hearts imaged at 96 hpf; arrow in (H) points to an extruding cardiomyocyte (n = 9/13 hearts). (I) klf2a+/-; klf2b-/- animals are more likely than WT siblings to exhibit cardiomyocyte extrusion upon Fgfr inhibition; number of treated larvae for each condition is shown above the individual columns. (J–K) Hearts of 96 hpf Tg(myl7: mCherry-CAAX); klf2 +/+ or klf2 -/- animals immunostained for pERK. (L–M) Hearts of 96 hpf Tg(fli1a:klf2b-p2A-tdTomato);Tg(myl7:EGFP-Hsa.HRAS); klf2 +/+ or klf2 -/- animals immunostained for pERK. Arrows and arrowheads point to extruding cardiomyocytes and pERK positive endocardial cells, respectively; V: ventricle, At: atrium; scale bars, 50 µm.
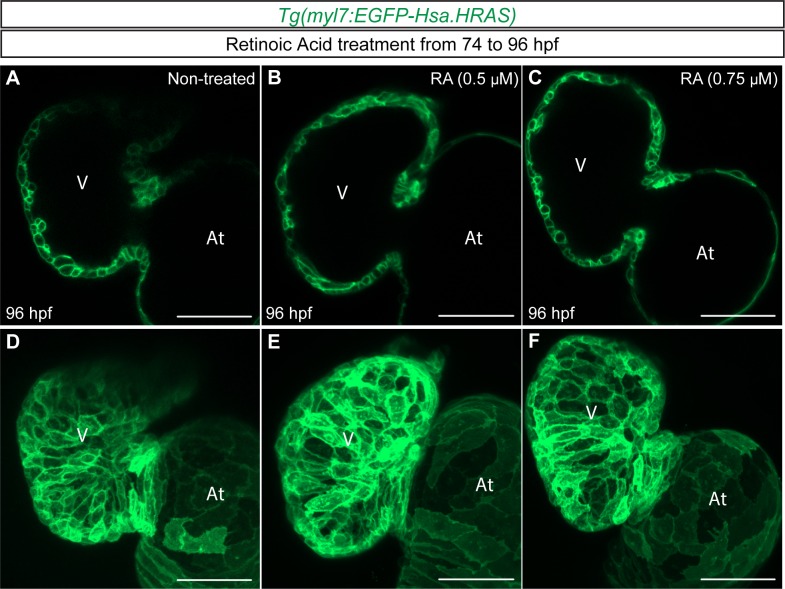
Figure 6—figure supplement 1. Increased Retinoic Acid signaling does not cause cardiomyocyte extrusion.
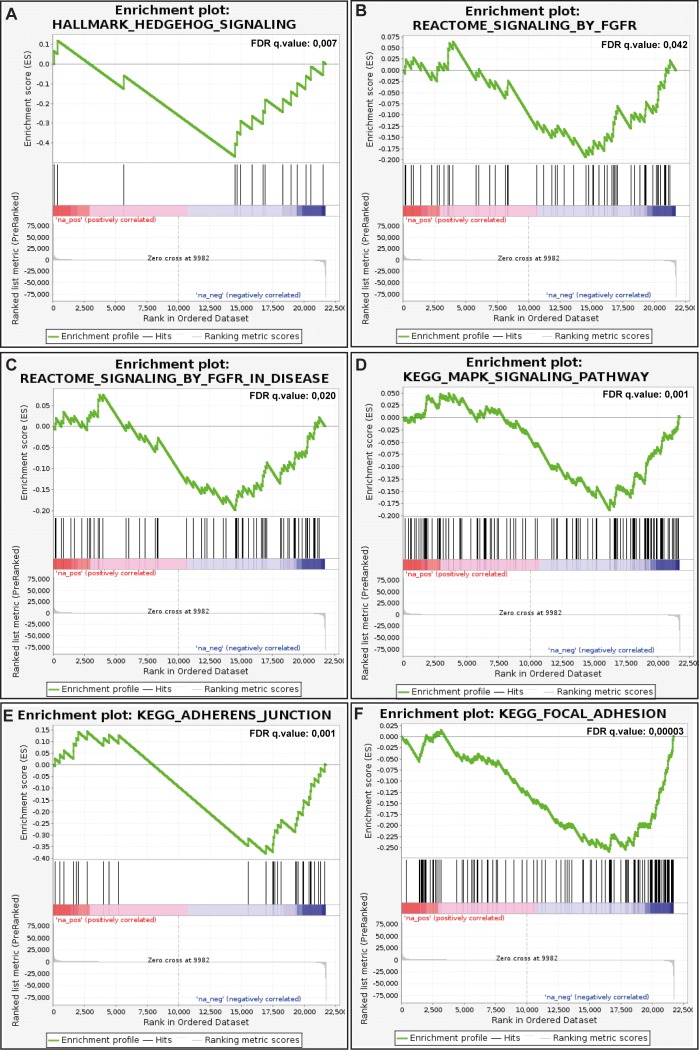
Figure 6—figure supplement 2. Broad GSEA enrichment plots of selected down-regulated gene sets.
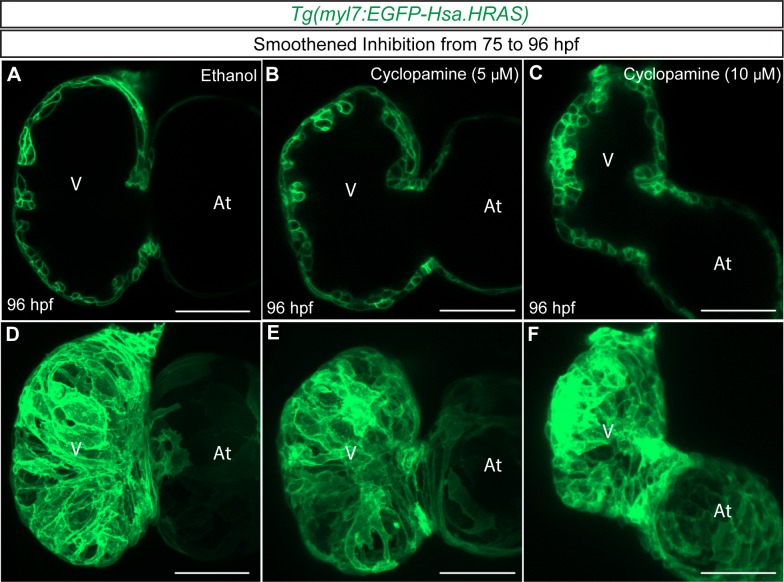
Figure 6—figure supplement 3. Inhibition of Hedgehog signaling does not cause cardiomyocyte extrusion.
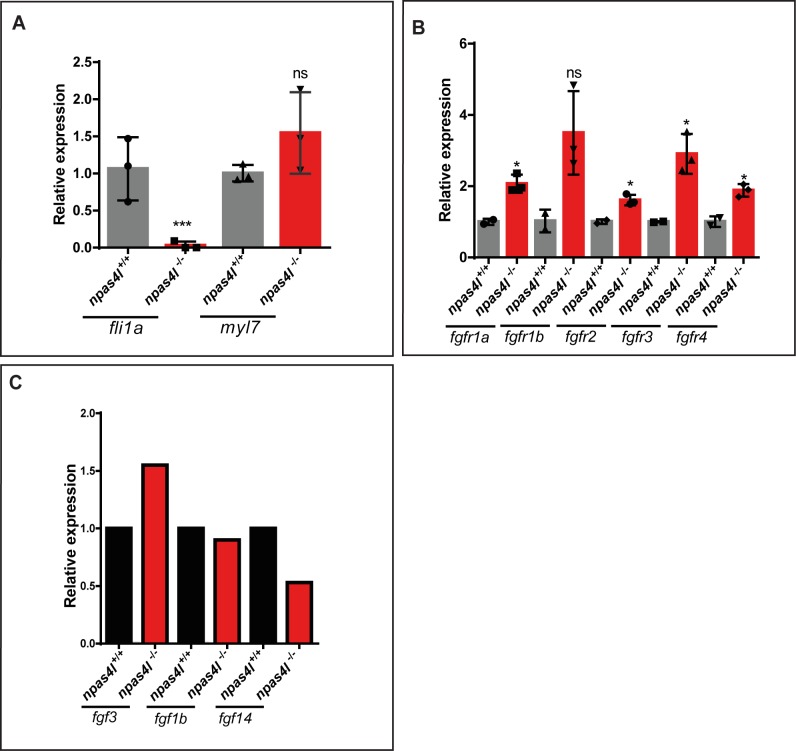
Figure 6—figure supplement 4. mRNA levels of fgf ligand and receptor genes in WT and klf2 mutant hearts.