Figure 3: Programming m6A-dependent transcriptional states with engineered reader modules.
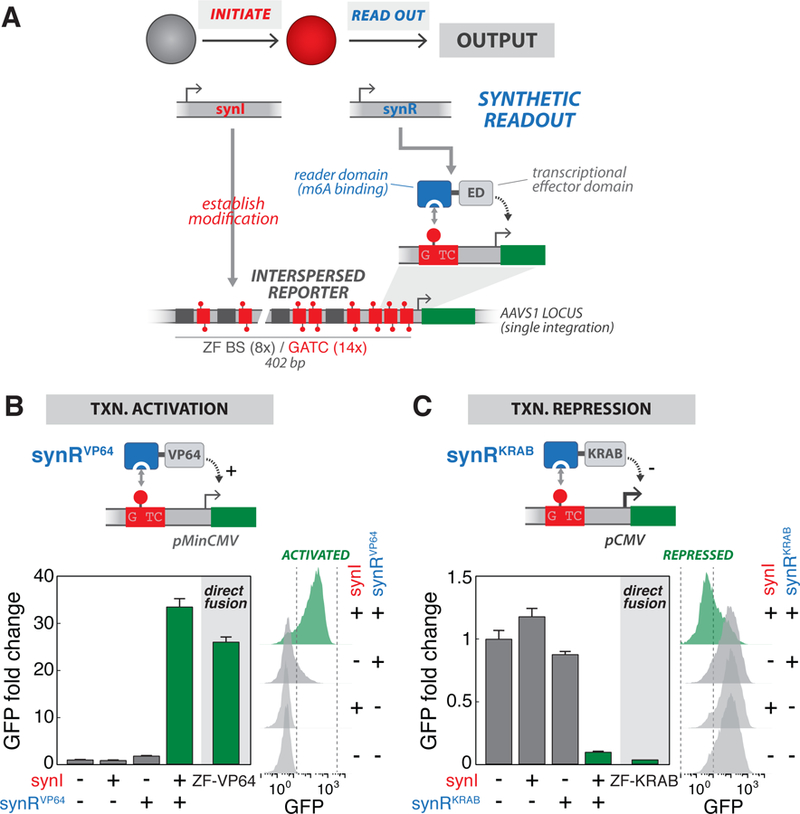
(A) Design of synthetic readout module (synR). synR is a fusion of an m6A “reader” domain (RD, binding domain of DpnI (aa146–254)) and a transcriptional effector domain (ED). m6A marks established by synI are specifically recognized by synR, which in turn regulates transcriptional activity of a reporter gene. For these experiments, we used stable cell lines harboring a singly-integrated Interspersed Reporter, with intermixed ZF BS and GATC sites upstream of a promoter (pMinCMV for activation or pCMV for repression), as the background strains., (B) Programming m6A-mediated transcriptional activation. Top: Schematic of the synRVP64 module, a fusion of DpnI m6A RD and VP64 transcriptional activation domain, which drives activation of a reporter gene via m6A recognition. Bottom: GFP fluorescence intensity, measured by flow cytometry, for cells transfected with indicated combinations of synI and synRVP64 expression constructs, or a direct ZF-VP64 fusion. Bottom left shows fold change of geometric mean GFP intensity normalized to the −/−condition (n=3; error bars, SD); bottom right shows raw flow cytometry distributions., (C) Programming m6A-mediated transcriptional repression. Top: Schematic of the synRKRAB module, a fusion of DpnI m6A RD and KRAB transcriptional repressive domain, which drives repression of a reporter gene via m6A recognition. Bottom: GFP fluorescence intensity, measured by flow cytometry, for cells transfected with indicated combinations of synI and synRKRAB expression constructs, or a direct ZF-KRAB fusion. See also Figure S3.
