Figure 4: Engineering spatial propagation of m6A with three-module read-write circuits.
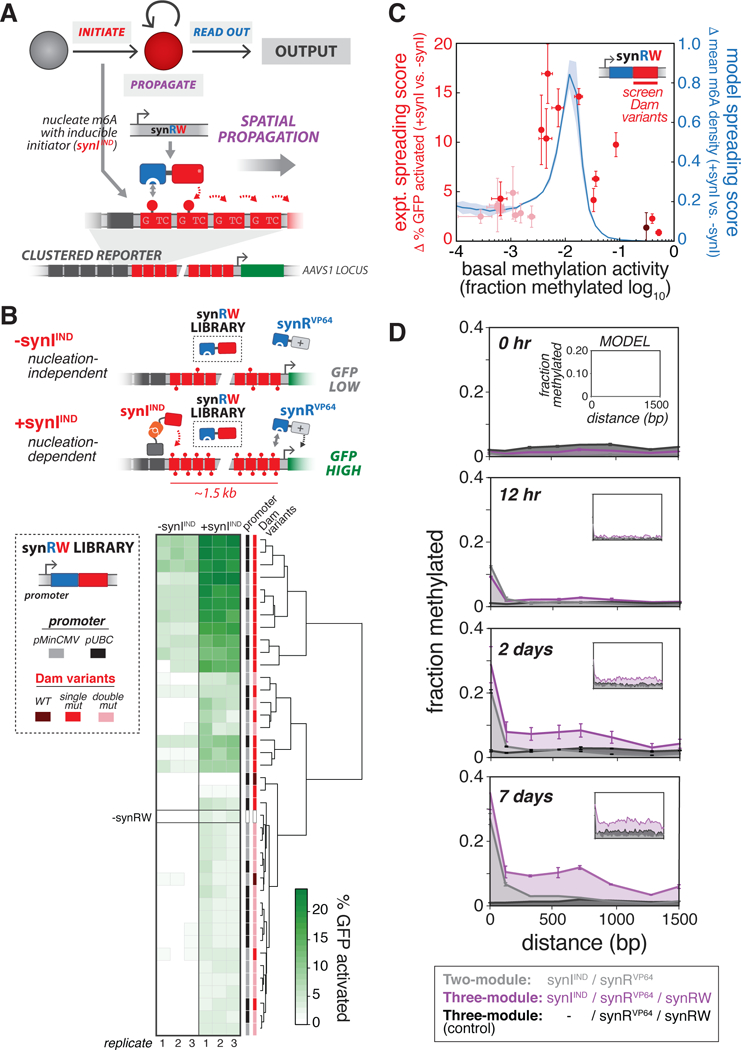
(A) Design of synthetic read-write module (synRW) for propagating m6A modifications over a domain. synRW is a fusion of DpnI m6A RD and a Dam writer domain (Dam variant). synRW recognizes pre-existing m6A marks and catalyzes methylation of nearby GATC motifs, creating local reinforcement and spreading of m6A to larger domains. For these experiments, we used Clustered Reporter cell lines, with pMinCMV driving expression of d2EGFP, as the background strain., (B) Screening synRW variants for nucleation-dependent spreading behavior. Top: Schematic of the “spatial propagation screen” design. A library of synRW variants was screened for the ability to activate a distal reporter gene in cells with synthetic initiator (+synIIND, GFP High), but not in cells lacking initiator (-synIIND, GFP Low). Bottom left: The synRW library featuring different promoter strengths and Dam mutant writers. Each member of the library was individually transfected into Clustered Reporter lines stably expressing synRVP64, either with (+synIIND) or without (-synIIND) stable expression of the inducible initiator. Bottom right: Screen results. Heat map of % GFP activated cells, quantified by flow cytometry 4 days following transfection of synRW (with continuous 200 μM ABA induction of synIIND). Hierarchical clustering analysis based on similarity in % GFP activated cells is shown. (C)Summary and modeling of spatial propagation screen. We defined quantitative metrics to score propagation propensity for synRW library members: “expt. spreading score” (red; n=3, error bars, SD) is the difference between % GFP activated cells with and without synIIND measured from the experimental screen; “model spreading score” (blue; mean ± SD) is the difference between model-computed m6A density at promoter-proximal sites with and without synI (see Methods, Figure S5). Shades of red correspond to Dam mutant writers: WT, single mutants, double mutants (dark to light, respectively)., (D) m6A profiles measured across the GATC array and over time for cells stably expressing the three-module “propagation circuit” (purple) or circuits lacking either synRW (grey) or synIIND (black). Cells were continuously induced with 200 μM ABA. Model simulations are shown in insets (with bZF =10, bDpnI =100, see Methods, Figure S5). (n=3; error bars, SD)., See also Figures S4-S6.
