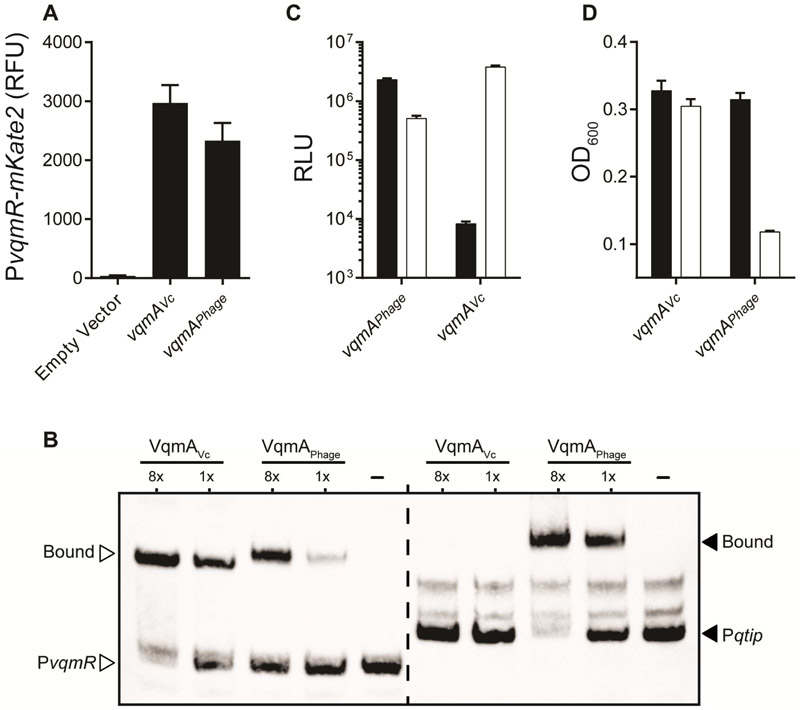
Figure 5: Asymmetric gene regulation by VqmAPhage and VqmAVc.
(A) Fluorescence from PvqmR-mKate2 in ΔvqmA V. cholerae carrying a vector control, vqmAVc under its native promoter, or arabinose inducible vqmAPhage. The medium contained 0.2% arabinose. RFU denotes relative mKate2 fluoresence units. (B) Electrophoretic mobility shift assay with PvqmR DNA (left, white arrowheads) and Pqtip DNA (right, black arrowheads) together with VqmAVc, VqmAPhage, or no protein (denoted -). Proteins used at 130 nM and 16.25 nM (denoted 8x and 1x, respectively). DNA probes = 540 pM. (C) E. coli carrying a plasmid with Pqtip-lux (black) or PvqmR-lux (white) and a second plasmid with either arabinose inducible vqmAPhage or vqmAVc. Medium contained 0.2% arabinose. We estimate that VqmAVc binds to its own (PvqmR) promoter 456-fold better than the non-cognate promoter (Pqtip), while VqmAPhage binds its target promoter (Pqtip) 4.5-fold better than the non-cognate promoter (PvqmR). PvqmR is activated 7-fold more strongly by the cognate VqmA (VqmAVc) than by the non-cognate VqmAPhage. In contrast, Pqtip is activated 278-fold more strongly by the cognate VqmA (VqmAPhage) than by the non-cognate VqmAVc. See Methods for details on quantitative comparisons. (D) Growth of ΔvqmA V. cholerae lysogenized with phage VP882 and either a vector carrying vqmAVc (left set of bars) or vqmAPhage (right set of bars). Black; no arabinose, white; 0.2% arabinose. In A, C, and D, data are represented as mean ± std with n=3 biological replicates. See also Figure S6.