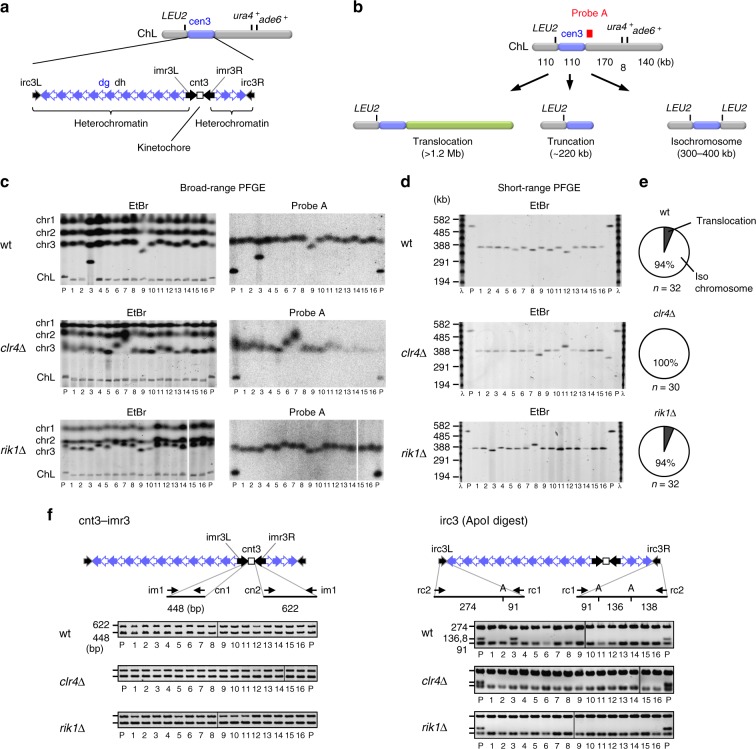
Fig. 2.
Clr4 and Rik1 suppress the formation of isochromosomes whose breakpoints are located in centromere repeats. a Repetitive sequences in cen3 of ChL are shown. Units of centromere repeats are indicated as arrows. b Illustration of the gross chromosomal rearrangement (GCR) products that have lost ura4+ and ade6+ from ChL: translocation, truncation, and isochromosome. The position of probe A used in Southern hybridization is indicated as filled box. c Chromosomal DNAs of wild-type, clr4∆, and rik1∆ strains (TNF5676, 5702, and 6121, respectively) were separated by broad-range pulse field gel electrophoresis (PFGE) and stained with ethidium bromide (EtBr). Positions of chr1, chr2, chr3, and ChL (5.7, 4.6, ~3.5, and 0.5 Mb, respectively) in the parental strain are indicated on the left of the panel. DNAs were transferred onto a nylon membrane and hybridized with probe A. P, Parental. d Chromosomal DNAs were separated by short-range PFGE and stained with EtBr. Sizes of the λ DNA ladder are indicated on the left of the panel. e Pie charts depict proportions of different types of GCRs. f Breakpoints were determined by PCR reactions using GCR products recovered from agarose gel. Both sides of cnt3–imr3 junctions were amplified in the reaction containing im1, cn1, and cn2 primers. irc3L and irc3R were amplified using rc1 and rc2 primers, and the PCR products were digested by ApoI and separated by agarose gel electrophoresis. A, ApoI. Uncropped images of depicted gels and blots are shown in Supplementary Figs. 10 and 11