Abstract
Auricularia auricula-judae is an edible mushroom and a traditional medicine in China as well as the fourth largest cultivated mushroom species in the world. Here for the first time, we present comparative transcriptome analyses of the fruiting bodies of three morphologically distinguishable A. auricula-judae cultivated varieties (Wujin, smooth; Banjin, partially wrinkled; and Quanjin, fully wrinkled) collected from Jilin Province, China. Biological triplicates were performed to determine the expression levels of 13,937 unigenes. Among them, only 13 unigenes were annotated to A. auricula-judae, highlighting the lack of publicly available reference sequences for this economically important species. Principal component analysis (PCA) determined that the gene expression profile of Quanjin was unique when compared to those of Banjin and Wujin. Such relationships were further supported by analyses of annotated and unannotated unigenes, differentially expressed unigenes, gene ontology functions, and the family of peroxidase genes. Using the KEGG database, significant alternations in biological pathways were detected among the three cultivars. This work contributes a large set of A. auricula-judae sequences to public database, establishes the relationships among major cultivars, and provides molecular guidance for breeding and cultivation.
Introduction
The Auricularia auricula-judae mushroom is commonly known as Jew’s ear, wood ear and jelly ear. The fruiting bodies are routinely consumed as an important edible mushroom in China. Furthermore, A. auricula-judae is also used as a traditional Chinese medicine with anti-tumor, detoxification, anticoagulant, hypoglycemic, and cholesterol-lowering properties1–3. In recent years, A. auricula-judae has been widely cultivated in large areas of Jilin, Heilongjiang, Henan and Hubei provinces in China, and the yield is ranked fourth in the world for mushroom production4,5.
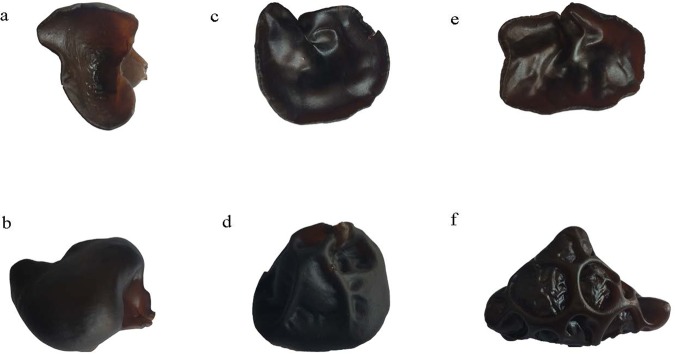
Based on the morphological characteristics of fruiting bodies, there are three major A. auricula-judae varieties in China: Wujin (smooth), Banjin (partially wrinkled), and Quanjin (fully wrinkled) (Fig. 1). The surface wrinkles (also known as veins) have been described as one of the major features of fruiting bodies6–8. The number and depth of wrinkles were deployed for cultivar identification purposes in China7. The physiological function of the wrinkles is to be determined although possibilities on reflecting nutrition and quality are widely implicated in the food market. All three varieties in China are believed to have been domesticated from wild origins. Quanjin originated from northern China, whereas Wujin and Banjin were more commonly cultivated in southern regions. Despite previous reports of genetic markers using analyses of inter-simple sequence repeat (ISSR), random amplification of polymorphic DNA (RAPD), and sequence related amplified polymorphism (SRAP)4,5,9–11, the molecular/genetic bases of differences between the three major Chinese varieties are largely unknown. Molecular studies can provide accurate identification and clarification of wild and domesticated varieties, thus facilitating breeding programs. However, neither the A. auricula-judae genome nor any transcriptome dataset is publicly available, representing a major barrier for molecular studies.
Figure 1.
Morphology of the three varieties of A. auricula-judae. Pictures show the apical and basal sides of the fruiting bodies of Wujin (Panels a and b), Banjin (Panels c and d), and Quanjin (Panels e and f).
Next-generation sequencing (NGS) technology is extensively used in life sciences for genome sequencing, transcriptome sequencing, and metagenomics sequencing12,13. Comparative transcriptome analysis is an effective way to compare gene expression patterns between different subjects and to provide insights into biological processes14,15. Moreover, transcriptome sequencing does not require a reference genome for data analysis13,16,17. Many transcriptome studies have been performed for mushroom species, including Lentius edodes18,19, Ganoderma lucidum20, Agrocybe aegerita21, Auricularia polytricha14, Pleurotus eryngii subsp. Tuoliensis15 and Cordyceps militaris22.
To determine the molecular relationships of the three major A. auricula-judae varieties in China, fruiting bodies from three cultivated strains (each represents a major variety) were examined for gene expression profiles using transcriptome sequencing: 13,937 unigenes were identified, and 12,813 were annotated. Principal component analysis (PCA) revealed that all three varieties had distinguishable gene expression profiles, but Quanjin was unique relative to the other two varieties. Further analysis of differentially expressed unigenes confirmed the PCA result. Such a molecular clarification of the major domesticated varieties provided directional information for developing breeding strategies. In addition, 1,124 unigenes did not display significant homology to any known genes, providing valuable information for further investigations into the biochemical and pharmaceutical properties of A. auricula-judae.
Results
De novo assembly of the A. auricula-judae transcriptome
Each of the individual RNA samples from Quanjin (n = 3), Banjin (n = 3), and Wujin (n = 3) (Fig. 1) were used to construct cDNA libraries for sequencing on an Illumina HiSeq2000 system. After removal of low quality reads, 69,814,486 reads, 62,918,023 reads, and 69,589,663 reads were obtained for Quanjin, Banjin, and Wujin, respectively (Supplementary Table S1). These high-quality reads were assembled de novo using Trinity23, and 33,316 contigs, 29,523 contigs, and 33,682 contigs were generated for Quanjin, Banjin and Wujin, respectively. To obtain a non-redundant library of consensus sequences, contigs of the three varieties were merged using TGICL24. Primary unigenes (54,320) were obtained containing 20,129 distinct clusters and 34,191 distinct singletons. The sequencing reads were further processed in edgeR25 to obtain FPKM (Fragments Per Kilobase of transcript per Million mapped fragments) values for each primary unigene in each of the biological samples (n = 9). For each variety, median FPKM value of the three replicates was used to represent the expression level. Only unigenes with a FPKM > 1.0 in any of the three varieties were collected for further analysis (Fig. 2). Ultimately, a final unigene library containing 13,937 unigenes (length range 201–30,448 bp) were established. The average and N50 lengths of unigenes were 2,594 and 3,521 bp, respectively (Supplementary Table S2).
Figure 2.
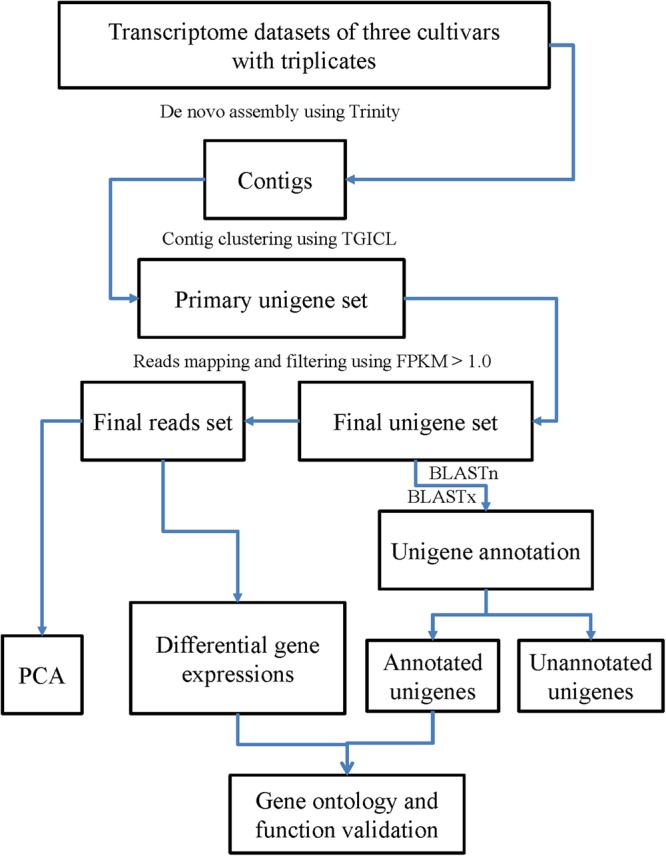
Analytical framework of the A. auricula-judae transcriptome. Raw reads of each of the three varieties were filtered and then de novo assembled into contigs by Trinity. Unigenes were obtained by clustering the contigs with TGICL. FPKM values were calculated for each of the unigenes, and only unigenes with FPKM >1.0 in any of the three varieties were used for further analyses and annotation against the public databases.
Annotation of unigenes
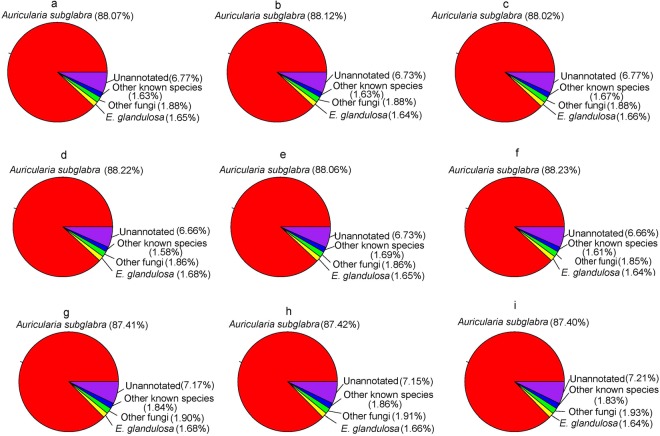
To identify if the unigenes were homologues of any known genes, a BLASTn search against the NCBI non-redundant nucleotide (nt) databases was performed and 12,011 unigenes were annotated. These unannotated unigenes were then searched against the Swiss-Prot and NCBI non-redundant protein (nr) databases using BLASTp with an E-value cutoff of 1e-4. Among the remaining 1,926 unigenes, 802 had significant similarities to known proteins. In total, 12,813 unigenes were annotated to known genes/proteins (Supplementary Dataset S1–S3). Only 0.0988% (median, n = 3), 0.101% (median, n = 3) and 0.101% (median, n = 3) of the unigenes were annotated to A. auricula-judae from varieties of Quanjin, Banjin and Wujin, respectively (Supplementary Table S3). Instead, the majority of unigenes were annotated to Auricularia subglabra (Fig. 3), highlighting lack of A. auricula-judae reference sequences in the public databases. In total, there were 1,124 unannotated unigenes among the nine samples (Supplementary Dataset S4).
Figure 3.
Summary of unigene annotation. Unigenes were divided into five major annotation groups (percentage >1% of total) and the proportion of each group is shown for each of the individual samples of Wujin (n = 3, Panels a–c), Banjin (n = 3, Panels d–f), and Quanjin (n = 3, Panels g–i).
Transcriptomic relationships between the three A. auricula-judae varieties
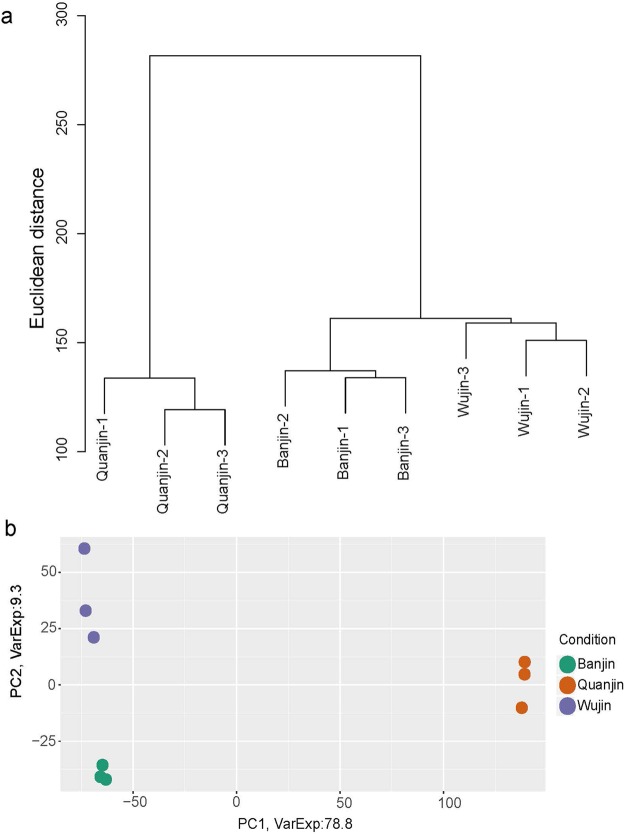
To determine the gene expression pattern relationships among the three varieties of A. auricula-judae, a cluster analysis using final reads set (Fig. 2) was performed. A Euclidean distance tree revealed that all three biological triplets clustered together in all varieties (Fig. 4a), confirming good sample quality. Quanjin (n = 3) was clearly isolated from Banjin (n = 3) and Wujin (n = 3), indicating an overall difference between their transcriptomic characteristics (Fig. 4a). PCA was also performed to confirm the relationships between the three varieties (Fig. 4b). The PCA revealed that Quanjin (n = 3) was clearly separated from Banjin (n = 3) and Wujin (n = 3) at both PC1 and PC2, whereas Banjin and Wujin appeared distinguishable only at PC2. At PC2, however, Quanjin (n = 3) was flanked by Banjin (n = 3) and Wujin (n = 3), indicating that Quanjin had more similarities to Banjin and Wujin than those shared between the latter two varieties in certain aspects (Fig. 4b). Therefore, the three mushroom varieties had a triangular relationship in their gene expression patterns. In addition to the between-variety relationships, the PCA results also revealed a visible difference in between-individual variations among the three varieties. Banjin displayed the least variation, whereas Wujin was the most variable, though the between-individual variations were mainly evident at PC2 (Fig. 4b).
Figure 4.
Gene expression relationships among the three A. auricula-judae varieties. (Panel a) Raw reads counts of unigenes in each of the samples were transformed into regularized log–transformed data by rlogTransformation. Then, the distance between samples was calculated and displayed in a hierarchical clustering tree by hclust. The X-axis labels samples, and the Y-axis represents the Euclidean distance between samples. (Panel b) The top two principal components (X-axis, PC1; Y-axis, PC2) deciphered 88.1% of all difference among the three varieties. Banjin and Wujin were similar to each other in PC1 but showed a greater difference in PC2 when compared to Quanjin.
Among the 12,813 unigenes annotated to known genes/proteins, the expression of 11,371 (88.7%) unigenes was shared in all three varieties (Fig. 5a). Quanjin uniquely expressed 637 annotated unigenes, whereas only 36 and 21 unigenes were uniquely detected in Banjin and Wujin, respectively. However, the latter two shared an intersection of 523 annotated unigenes whose expression was not detected in Quanjin (Fig. 5a). Between these sets of 637 Quanjin unique unigenes and 523 Banjin-Wujin shared unigenes, 141 pairs of unigenes had nucleotide sequence similarities (determined by local BLASTn) and 57 pairs of unigenes had amino acid sequence similarities (determined by local BLASTx), suggesting that they may perform similar functions but have polymorphisms in the sequences. Taken together, the above results indicated that Banjin and Wujin transcriptomes are closely related to each other, while they were distant to Quanjin in the gene expression landscape.
Figure 5.
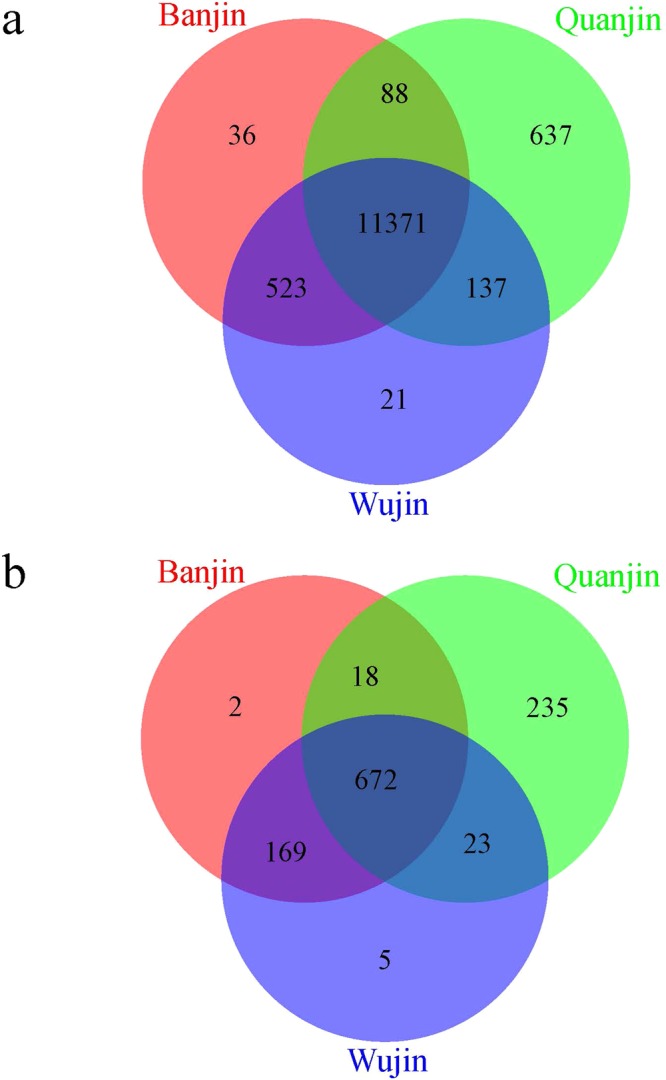
Unigene expression. The numbers of unigenes detected in the three A. auricula-judae varieties are displayed in Venn charts. (Panel a) Expression of unigenes annotated to Auricularia subglabra, E. glandulosa, and other species (Fig. 3). (Panel b) Expressions of unannotated unigenes (Fig. 3).
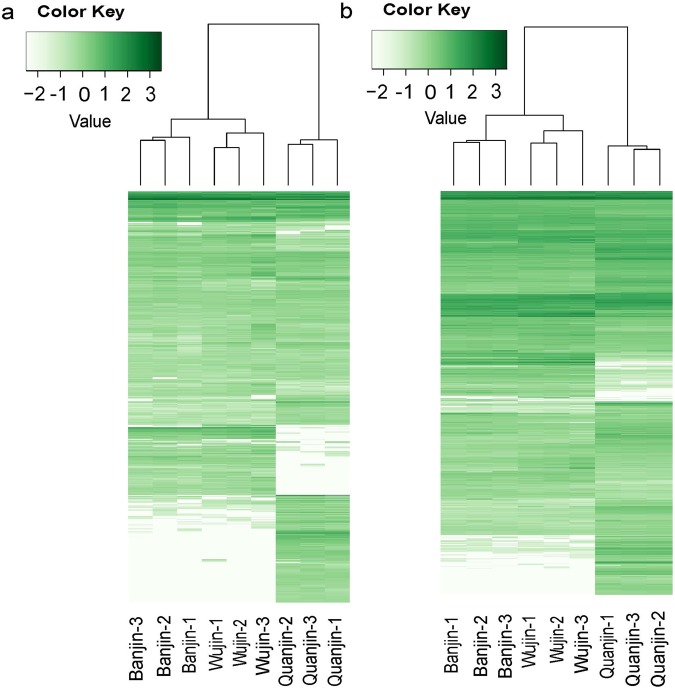
Among the 1,124 unannotated unigenes, 672 (59.8%) were shared by all of the three A. auricula-judae varieties. Quanjin uniquely expressed 235 novel unigenes, whereas 169 novel unigenes were only detected in the intersection between Banjin and Wujin (Fig. 5b). Thirty pairs of homologues were detected in these two sets of novel unigenes by local BLAST. The expression levels of these novel unigenes in all nine samples are displayed in a clustering heat map using the FKPM values (Fig. 6a). The expression patterns of the novel unigenes (Fig. 6a) conformed to the whole transcriptome characteristics and supported that Quanjin was isolated from Banjin and Wujin (Fig. 4).
Figure 6.
Expression profiles of novel and differentially expressed unigenes. The FPKM of all A. auricula-judae unigenes in nine samples were scaled together and transformed into Z-scores ((x-mean)/(s.d.)) using an in-house Perl program and are represented in a clustering heat map by Heatmap 2. The Z-scores are color coded as shown in the color bar. (Panel a) Expression profile of novel unigenes. (Panel b) Expression profile of differentially expressed unigenes.
Differentially expressed unigenes
Differentially expressed unigenes (DEGs) were determined for each of the two variety pairs using EdgeR with a cutoff threshold of P < 0.05. There were 2,861 DEGs in Banjin vs. Wujin, 5,992 DEGs in Quanjin vs. Banjin, and 6,549 DEGs in Quanjin vs. Wujin (Supplementary Table S4). An expression clustering heat map of all identified DEGs was produced for all nine samples using the FKPM values (Fig. 6b). The DEG clustering results also conformed to the whole transcriptome clarification that Quanjin was more divergent compared to the other two A. auricula-judae varieties (Fig. 4).
Gene ontology (GO) annotation and cluster analysis of function groups
The 12,813 BLAST-annotated A. auricula-judae unigenes were further annotated to 162 GO functions. Among them, 55 GO functions belonged to the category of molecular function, 50 belonged to cellular component, and 57 belonged to biological process (Fig. 7, Supplementary Dataset S5). Based on the number of unigenes in each GO function detected in each of the three varieties, a GO function clustering analysis was performed to summarize the relationships among the three varieties at the GO function level. The numbers of unigenes were transformed together into Z-scores and displayed separately as DEGs and un-DEGs (Fig. 7). Again, Banjin and Wujin displayed greater similarities than those of Banjin vs. Quanjin and Wujin vs. Quanjin. This is shown in Fig. 7 in two aspects as the greatest number of un-DEGs and the lowest numbers of DEGs between Banjin and Wujin (Fig. 7 and Supplementary Dataset S5), indicating similarities between the two varieties. There was further supporting evidence from the DEGs section, where the columns of up-regulated Quanjin unigenes clustered together (bq-q and qw-q, Fig. 7) and was isolated from the cluster containing columns of down-regulated Quanjin unigenes (bq-b and qw-w, Fig. 7), indicating similar patterns for the difference between Qianjin vs. Banjin and Qianjin vs. Wujin. Based on these results, the functional characteristics of Quanjin transcriptome were indeed unique compared to those of Banjin and Wujin.
Figure 7.
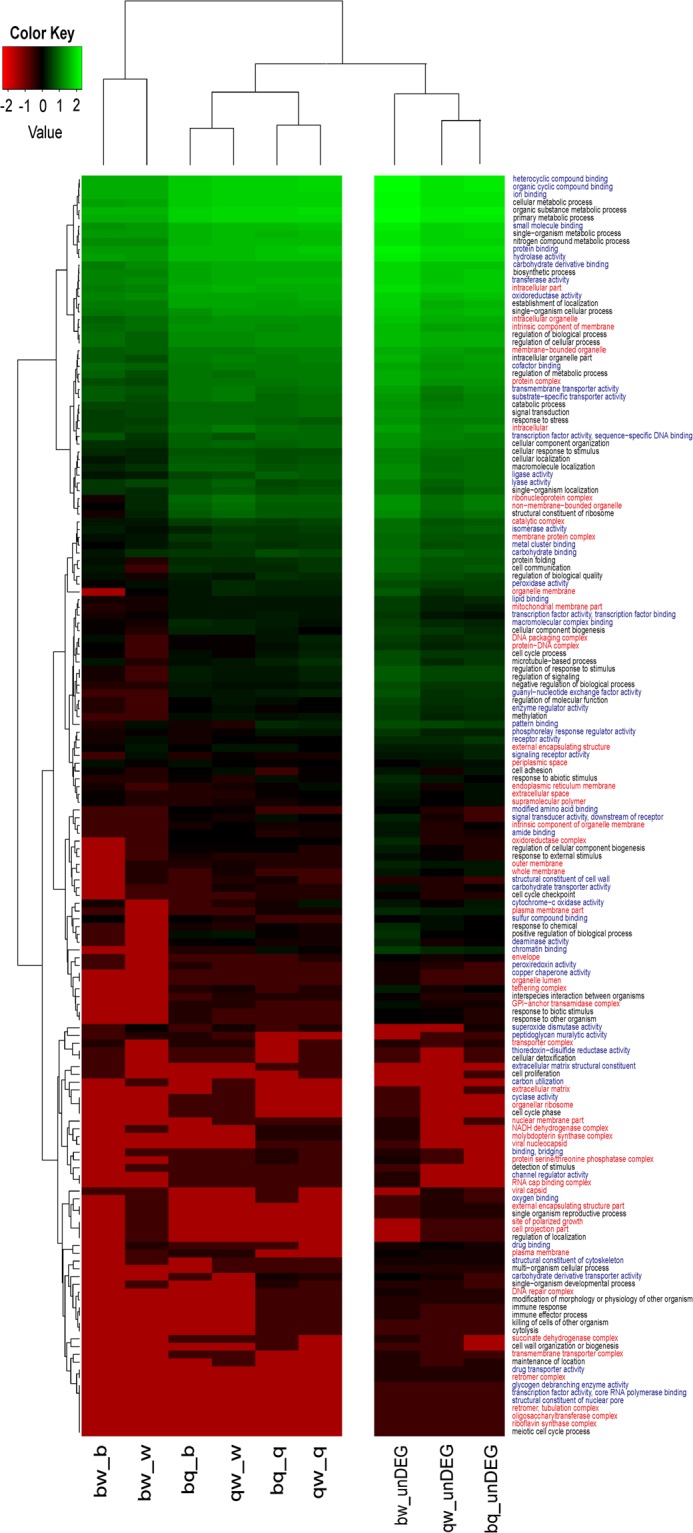
GO of DEGs and unDEGs. The numbers of unigenes in each GO term were collected for DEGs (P < 0.05) and unDEGs, respectively, for each pairwise comparison among the three varieties. All of the unigene numbers were normalized into Z-scores ((x-mean)/(s.d.)) using an in-house Perl program and are represented in a clustering heat map by Heatmap 2. The Z-scores are color coded as shown in the color bar. Color word labeling was used to represent different classes of GO categories of molecular function (blue), cellular component (red), and biological process (black).
KEGG pathway enrichment associated with surface wrinkle
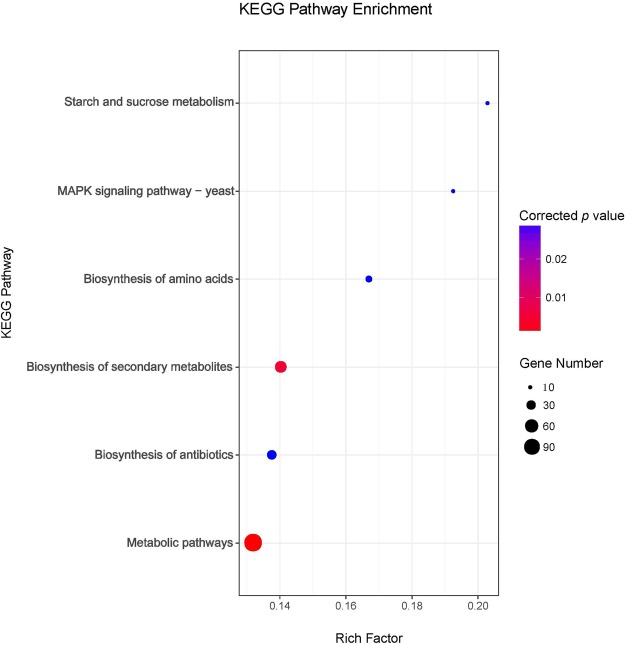
983 unigenes were extracted for wrinkle down-regulation (FPKM: Wujin > Banjin > Quanjin) and 988 unigenes were extracted for wrinkle up-regulation (FPKM: Wujin < Banjin < Quanjin). The total of 1971 unigenes were pooled and determined for significant enrichments (with corrected P < 0.05, by Benjamini-Hochberg FDR method) in six KEGG pathways, such as starch and sucrose metabolism (Rich Factor, number of enriched genes/number of total genes in the pathway, RF = 0.20), MAPK signaling pathway-yeast (RF = 0.19), biosynthesis of amino acids (RF = 0.17), biosynthesis of secondary metabolites (RF = 0.14), biosynthesis of antibiotics (RF = 0.14) and metabolic pathways (RF = 0.13) (Fig. 8, Supplementary Dataset S6). To our knowledge, this is the first description of biological functions possibly associated with fruiting body wrinkles, a major morphological feature of Auricularia mushrooms6–8.
Figure 8.
KEGG pathways associated with fruiting body wrinkle Scatter plot of KEGG pathways enriched for unigenes that were up- or down-regulated by the wrinkle morphology. The X-axis displays the Rich Factor which is the ratio of numbers of the unigenes in a pathway term to all gene numbers annotated in the same pathway term. The size of bubble represents the number of unigenes and the color gradient indicates the corrected P value by Benjamini-Hochberg FDR method. Only pathways with corrected P < 0.05 were shown.
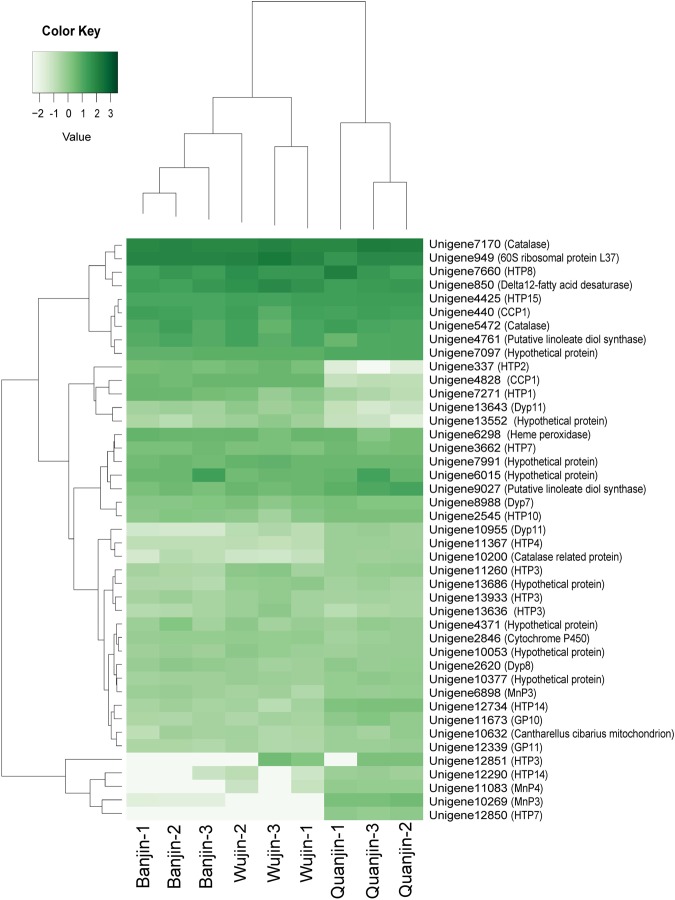
Expressions of peroxidase-like unigenes
To further validate the relationship of functional characteristics among the three A. auricula-judae varieties, we examined the expression patterns of peroxidase-like unigenes in all nine samples. In total, 43 unigenes displayed homologies to known peroxidases (Supplementary Dataset S7), including 14 heme-thiolate peroxidase (HTP)-like unigenes, four DyP-type peroxidase-like unigenes, and three manganese peroxidase (MnP)-like unigenes (Supplementary Figure S1). Figure 9 shows a clustering heat map based on the FKPM values. Consistent with previous results (Figs 4, 6 and 7), Quanjin (n = 3) was isolated from Banjin (n = 3) and Wujin (n = 3) samples, confirming that the expression profiles of peroxidase-like genes in Quanjin were unique when compared to those of Banjin and Wujin (Fig. 9). However, from the peroxidase expression profiles, clustering analysis could not sufficiently separate Banjin and Wujin, mainly due to a large between-individual variation in Wujin (Fig. 9, Wujin-2). This result conformed to the PCA results that the greatest between-individual variation was observed in Wujin when all unigenes were used (Fig. 4b).
Figure 9.
Expression profiles of peroxidase-related unigenes. The Z-scores ((x-mean)/(s.d.)) of FPKM values of A. auricula-judae peroxidase-related unigenes were extracted from the Z-scores of all unigenes and are represented in a clustering heat map by Heatmap 2. The Z-scores are color coded as shown in the color bar. Forty-three unigenes were functionally annotated to peroxidase by GO and were labelled using the unigene ID followed by the BLASTn/x annotations (Supplementary Dataset S7).
Real time PCR validation of selected peroxidase-like unigene transcriptions
To validate the transcriptome results, 16 peroxidase-like unigenes (Supplementary Figure S1) were selected for RT-qPCR analysis. Firstly, Sanger sequencing of the RT-qPCR products (Supplementary Dataset S8) confirmed genuine amplifications of all of the 16 peroxidase-like transcripts tested (Supplementary Dataset S9). Secondly, the ΔCt values had negative correlations (P < 0.05) to the FKPM values in all three cultivars (Supplementary Table S5), showing compatibility of expression levels determined by the two methods. Interestingly, transcriptions of A. aj DyP1 (JQ650250)26 were detected by RT-qPCR with relatively high ΔCt values (Supplementary Table S6) in all three cultivars although its homologue was not included in our final unigene set of FPKM > 1.0. Further investigation using contigs detected signals of two non-overlapping JQ650250 homologues with low FPKM values (Supplementary Table S6), emphasizing that the final unigene set excluded lowly expressed A. auricula-judae genes.
Discussion
In 2012, the genome of Auricularia subglabra was sequenced and annotated27. The transcriptome of A. polytricha was sequenced using the Illumina RNA-seq protocol, and a gene expression profile was reported in 201414. Recent studies of A. auricula-judae, an important edible mushroom in China, however, are largely limited in classification, phylogenetic analysis, biodiversity, and conservation issues using ISSR, RAPD, and SRAP5,9–11. Knowledge of the genomics and transcriptomics of A. auricula-judae is needed to facilitate research activities and agricultural developments. To address a fundamental question concerning the gene expression characteristics of three major Chinese domesticated A. auricula-judae varieties, we applied comparative transcriptome analysis to the fruiting bodies of Quanjin, Banjin and Wujin. A clear relationship was determined that Quanjin displayed major differences in gene expression compared to Banjin and Wujin (Fig. 4b, PC1), conforming to a previous report that A. auricula-judae strains with different fruiting body wrinkle characteristics belonged to two major types based on SRAP classification28. This observation was further supported by expression patterns of both annotated unigenes (Fig. 7) and unannotated novel unigenes (Fig. 6a), as well as DEGs and unDEGs (Fig. 7). Functional validations also supported the conclusion by GO function categorization (Fig. 7) and the expression of peroxidase-like genes (Fig. 9). This conclusion supports the notion that Wujin and Banjin may have a genetic relationship or common ancestor from southern China, but that Quanjin had its own origin from northern China. Genomic sequence data would be needed to reveal the full picture of genetic relationship29. Encouragingly, during reviewing period of this manuscript, genomes of 90 mushroom species were published30. From a genome of A. heimuer, 16,244 putative genes were predicted31, comparable with the number of unigenes reported in this study.
The dye de-colourizing peroxidase (DyPs) are a family of heme peroxidases that are considered possible resources for the bioremediation of lignin. DyPs have been isolated from A. auricula-judae with unique sequence and enzymatic features26. We detected the expression of four previously unreported DyP-like unigenes based on their sequence homology to Auricularia subglabra DyPs (Supplementary Figure S1b). Expression of the previously reported A. auricula-judae DyP (A. aj-DyP1 derived from Strain DSMZ 11326, NCBI accession number JQ650250, Supplementary Figure S1b), however, was not detected by the transcriptome final unigene set (Supplementary Datasets S1–S3) but detected by RT-qPCR (Supplementary Table S6). Further investigation on the assembly result revealed two non-overlapping contigs that displayed more than 99% identity to JQ650250 but had low expression levels (FPKM < 0.3), emphasizing that our final unigene set excluded lowly expressed genes due to the filtration of FPKM > 1.0. The low expression levels of A. aj-DyP1 may be due to possible genotypic differences between the three Chinese varieties we used and Strain DSMZ 11326 from which A. aj-DyP1 was discovered. It may also be due to phenotypic factors that can be affected by environmental and/or cultivation conditions. Nevertheless, the diversities of A. aj-DyPs (Fig. 9, Supplementary Fig. S1b) are an excellent example of the high degrees of variation in A. auricular-judae gene expression. Clarifications of genetic polymorphism and phenotypic plasticity will be challenging tasks for the A. auricular-judae research and horticulture industries.
Six KEGG pathways were identified for significant relationships to the fruiting body wrinkle characteristics (Fig. 8, Supplementary Dataset S6). This result strongly suggested that wrinkle characteristics may not be controlled by a single gene. The presence of pathways of starch and sucrose metabolism, biosynthesis of amino acids and biosynthesis of secondary metabolites strongly supported the notion that food quality may vary among the cultivars with different wrinkle characteristics6–8. Biosynthesis of secondary metabolites is also important for the production and accumulation of pharmaceutical compounds, suggesting that different cultivars may have specific values of pharmaceutical development. Involvement of the biosynthesis of antibiotics pathway suggested difference in traits of disease resistance, therefore implicating potentials in breeding strategies.
In this study, an A. auricular-judae sequence library containing 13,937 unigenes was established and made publicly available (as a transcriptome shotgun assembly project that was deposited at DDBJ/EMBL/G under the accession GFZV00000000; see Supplementary Dataset S4 for the unannotated unigenes). Previously, only 13 A. auricular-judae gene sequences in our library had been available in public databases (Supplementary Table S3). Therefore, this work made a significantly knowledge advance for identifying A. auricular-judae gene expressions. The vast majority of these genes could be annotated with known GO functions, providing valuable information for understanding specific functions in future studies. Among the 1,124 unannotated unigenes that did not have significant sequencing similarities to known genes/proteins, expression of 627 unigenes was detected in all of the three varieties (Fig. 5b). These commonly expressed novel genes are likely to play roles in A. auricular-judae specific functions. In addition to its nutritional value in the food industry and novel enzymatic activities for the interests of biotechnology developments, A. auricular-judae fruiting bodies also produce bioactive molecules with pharmaceutical properties1–3, including anti-oxidant activities32. One of the particular encouraging areas is for anti-tumor activities33–36. Polysaccharides from A. auricula-judae inhibit the proliferation of Acinar cell carcinoma and induce apoptosis in S-180 tumor cells by upregulating the expression of Bax gene and down-regulating the Bcl-2 expression37. A. auricula polysaccharides also exhibit antiproliferative effects in HepG2 and Bel-7404 human hepatoma cells by inducing apoptosis and G1 or S cell cycle arrest38. Anti-proliferative activities, including high lactate dehydrogenase (LDH) levels, have been detected against human acute myelocytic leukemia U937 cells from A. auricula-judae extracts39. The novel genes, particularly those commonly expressed in all varieties (Fig. 4b), could be potential subjects for investigating anti-tumor activities.
To conclude, by analyzing the expression patterns of 13,937 A. auricula-judae unigenes, we determined the relationships among the three major cultivars in China. Supported by the expression patterns of all genes, annotated genes, un-annotated genes, differentially expressed genes and peroxidase genes, Quanjin displayed unique characteristics to Banjin and Wujin at the levels from specific gene function to global gene expression, indicating that the morphological difference of wrinkles in fruiting body associated with activities of many genes. Determination of altered activities in certain biological pathways associated with the wrinkle characteristics strongly suggested that fruiting bodies of the three cultivars contained different combinations of bioactive substances, supporting the implication that wrinkle characteristics may reflect nutritional and pharmaceutical properties.
Methods
Sampling, RNA extraction, and deep sequencing
Three varieties of A. auricula-judae known as Quanjin (Strain Hei29), Banjin (Strain Heibao), and Wujin (Strain Heishan) (Fig. 1) were collected from the “Huangsongdian National Station for A. auricula-judae Cultivation, Standardization and Demonstration”, Jiaohe City, Jilin Province, China, during the harvest season of autumn 2015. Individual fruiting bodies of each variety were sampled in triplicate, frozen in liquid nitrogen, and then stored at −80 °C prior to RNA extraction.
Total RNAs were isolated with TRIzol reagent (Invitrogen, USA) according to manufacturer’s protocols. The quality and quantity of RNA extracts were assessed using agarose gel electrophoresis and a Nanodrop 2000 Spectrophotometer (Thermo Scientific, USA). More than 5 µg of total RNA was obtained from each of the individual samples with OD260/280 > 1.8 and OD260/230 > 1.8.
The cDNA libraries were constructed for each individual sample using an Illumina TruSeq Stranded mRNA Library Preparation Kit LT for mRNA (Illumina, USA). The libraries were then individually sequenced using the manufacturer’s 125-nt pair-end protocol of the Illumina HiSeq platform at LC Sciences (China). Raw reads were filtered, and adapter sequences were removed by the manufacture’s quality control pipeline. The dataset has been deposited in NCBI/SRA database with accession numbers of SRX3295820 (Quanjin), SRX3295819 (Banjin), and SRX3304706 (Wujin).
De novo assembly and unigene library
High quality reads were pooled for each of the varieties for de novo assembly using Trinity23. The resulting three contig libraries were clustered with TGICL-2.124 by parameters of “-l 40 -v 20” to form a primary unigene library. It is worth noting that the unigene library does not represent polymorphisms although this method has been reported previously for comparative transcriptome analysis at intra-species level40–47.
All clean reads of each individual sample were mapped to the primary unigene library by bowtie-1.1.248, respectively, with the following parameters “-q–phred33-quals -n 2 -e 99999999 -l 25 -I 1 -X 1000 -p 10 -a -m 200”. Then, the mapping output of each sample was calculated for read counts for each unigene by HTSeq-0.6.1p149. The HTSeq outputs were further processed in edgeR25,50 to obtain the FPKM value for each unigene in each of the individual samples. For each variety, the median FPKM value of the three replicates was used to represent the expression level. To minimize possible experimental error, an expression threshold of FPKM > 1.0 was set to filter out weak expression. The unigenes with a median FPKM > 1.0 in any of the three varieties were used to form the final unigene library (Fig. 2).
PCA
Raw read counts of the final unigene library were transformed to regularized log–transformed data by rlogTransformation51 to avoid domination of highly variable genes and to have a roughly equal contribution from all genes during the distance measuring. Gene expression cluster analysis was performed by heatmap.2. PCA was performed by the function prcomp of R. The principal component scores were plotted by qplot function of ggplot2 package.
Annotation of unigenes
The NCBI-nt, NCBI-nr, and Swiss-prot databases were used to annotate the final A. auricula-judae unigene library. First, unigenes were searched against NCBI-nt by BLASTn, and the best hit (e-value < 1e-5) were retained as the nucleotide annotation. Second, the remaining unannotated unigenes were searched against the NCBI-nr database by BLASTx, and the best hits (e-value < 1e-4, identity > 30%) were selected as the protein annotation. Finally, the remaining unannotated unigenes were aligned to the Swiss-prot database using BLASTx with a cut-off threshold of e-value < 1e-4 and identity > 30%. The unigene sequences were deposited at DDBJ/EMBL/G under the accession number GFZV00000000.
DEGs and GO functions
Based on the read counts, DEGs were identified using EdgeR (P < 0.05). Pairwise comparisons were performed among the three varieties. For each of the pairwise comparison, DEGs and unDEGs were separately recorded and used as inputs for GO annotations using Interproscan-5.2–45.052 with the following parameter “ -t n -f tsv -goterms -pa -iprlookup -dp”. The GO annotations were collected. The number of unigenes for each GO term on Level-3 was calculated for the DEGs and unDEGs, respectively, by an in-house Perl program. The number of unigenes for a GO term was then normalized as Z-score ((x-mean)/(s.d.)) and represented in a heat map using an in-house Perl program.
KEGG pathway enrichment associated with surface wrinkle
To identify genes that possibly associate with the fruiting body wrinkle characteristics, the transcriptome data (FPKM) for each unigene was ranked for the order of Wujin, Banjin and Quanjin, using the Short Time series Expression Miner (STEM) program53. Two sets of unigenes were extracted using FPKM values of Wujin > Banjin > Quanjin (wrinkle down-regulation) or Wujin < Banjin < Quanjin (wrinkle up-regulation). These two sets of unigenes were pooled then examined in KOBAS 3.054 to identify gene enrichments in KEGG pathways, with default parameters and using Auricularia delicata as the reference species.
Real time PCR validation of selected peroxidase-like unigene transcriptions
DNA primers of 16 selected peroxidase unigene targets and the A. aj-DyP1 (NCBI Accession Number: JQ650250, Supplementary Figure S1) were designed using Primer Primer 5.0 software (http://www.premierbiosoft.com). Total RNA concentration was measured with Nanodrop 2000 (Thermo Fisher, USA) for each sample and 1 µg of RNA was used from each sample to synthesize the cDNA by the PrimeScript RT Master Mix kit (TaKaRa, Japan). Subsequently, quantitative (q)PCR was performed using the SYBR Premix Ex Taq kit (Tli RNaseH Plus) (TaKaRa, Japan) on a QuantStudioTM 7 Flex Real-Time PCR Detection System (Applied Biosystems, USA) following the manufacturer’s instructions. Each reaction was performed in a total volume of 20 µL with 1 µL of undiluted cDNA, 10 µL TB Green Premix Ex Taq II, 0.4 µL ROX Reference Dye II (SYBR Green, TaKaRa, Japan), 0.8 µL forward and reverse primer (10 µM), respectively. The PCR program was 95 °C for 10 min, 40 cycles (95 °C for 15 s, 60 °C for 30 s) with a single melting curve fluorescence measurement (60 °C–95 °C, +0.05 °C/s). Each experiment was performed in triplets with ddH2O replacing the cDNA as negative control. The Ct numbers were recorded and the qPCR results were analyzed using the ΔCt method using the 18 S rRNA as the reference gene55. Pearson correlation analysis was performed between ΔCt (median) and FKPM (median) values using MiniTab. All the RT-qPCR products of each cultivar were pooled for the agarose gel electrophoresis followed by gel excision and DNA purification using Gel Purification Kit (TIANGEN, China). The purified PCR products were sent for Sanger sequencing by GENEWIZ Corporation (SuZhou, China). The resulting DNA sequences were used to verify genuine amplifications of the target unigene fragments using local Blastn against the target unigene sequences.
Supplementary information
Acknowledgements
This work was supported by the National Key R&D Program of China (2016YFC1302100); the National Natural Science Foundation of China (81472827 and 81773023); and the Hundred-Talent Program and Frontier Research Program (QYZDB-SSW-SMC038) of the Chinese Academy of Sciences.
Author Contributions
Y.G.T., S.G. and H.W. designed and supervised the study. D.S.Z., R.L., T.Y.C., Y.B.Z. and X.J.L. conducted the experiments, and Y.H.Z., L.W., H.W. and S.G. analyzed data. Y.H.Z., H.W., L.W., D.S.Z. and S.G. wrote the paper. G.W. revised the paper. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yuhui Zhao and Liang Wang contributed equally.
Contributor Information
Hui Wang, Email: huiwang789@gmail.com.
Shan Gao, Email: gaos@sibet.ac.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36984-y.
References
- 1.Ma H, Xu X, Feng L. Responses of antioxidant defenses and membrane damage to drought stress in fruit bodies of Auricularia auricula-judae. World J Microbiol Biotechnol. 2014;30:119–124. doi: 10.1007/s11274-013-1416-z. [DOI] [PubMed] [Google Scholar]
- 2.Reza MA, et al. Dichlormethane Extract of the Jelly Ear Mushroom Auricularia auricula-judae (Higher Basidiomycetes) Inhibits Tumor Cell Growth In Vitro. International Journal of Medicinal Mushrooms. 2014;16:37–47. doi: 10.1615/IntJMedMushr.v16.i1.40. [DOI] [PubMed] [Google Scholar]
- 3.Dai YC, Yang ZL, Cui BK, Yu CJ, Zhou LW. Species Diversity and Utilization of Medicinal Mushrooms and Fungi in China (Review) International Journal of Medicinal Mushrooms. 2009;11:287–302. doi: 10.1615/IntJMedMushr.v11.i3.80. [DOI] [Google Scholar]
- 4.Du P, Cui BK, Zhang CF, Dai YC. Genetic diversity of wild Auricularia auricula-judae revealed by ISSR analysis. Biochemical Systematics and Ecology. 2013;48:199–205. doi: 10.1016/j.bse.2012.11.011. [DOI] [Google Scholar]
- 5.Yan P-S, Luo X-C, Zhou Q. RAPD molecular differentiation of the cultivated strains of the jelly mushrooms, Auricularia auricula and A. polytricha. World Journal of Microbiology & Biotechnology. 2004;20:795–799. doi: 10.1007/s11274-004-5840-y. [DOI] [Google Scholar]
- 6.Barrett MF. Three Common Species of Auricularia. Mycologia. 1910;2:12–18. doi: 10.1080/00275514.1910.12020751. [DOI] [Google Scholar]
- 7.Li L, Fan XZ, Liu W, Xiao Y, Bian YB. Comparative analysis on the diversity of Auricularia auricula-judae by physiological characteristics, somatic incompatibility and TRAP fingerprinting. World Journal of Microbiology & Biotechnology. 2011;27:2081–2093. doi: 10.1007/s11274-011-0671-0. [DOI] [Google Scholar]
- 8.Priya RU, Geetha D, Darshan S. Biology and Cultivation of Black Ear Mushroom – Auricularia spp. Advances in Life Sciences. 2016;5:10252–10254. [Google Scholar]
- 9.Zhang JC, Ma QF, Zhang PQ, Dai XD. Identification of cultivated strains of Auricularia auricula from northeastern China by ISSR marker. Myoyma. 2007;4:534–538. [Google Scholar]
- 10.Tang L, Xiao Y, Li L, Guo Q, Bian Y. Analysis of Genetic Diversity Among Chinese Auricularia auricula Cultivars Using Combined ISSR and SRAP Markers. Current Microbiology. 2010;61:132–140. doi: 10.1007/s00284-010-9587-4. [DOI] [PubMed] [Google Scholar]
- 11.Tao P, Xu X, Liu H. Genetic Diversity Analysis of Wild Auricularia auricul from Heilongjiang Province. Chinese Agricultural Science Bulletin. 2011;27:182–186. [Google Scholar]
- 12.Maclean D, Jones JD, Studholme DJ. Application of ‘next-generation’ sequencing technologies to microbial genetics. Nature Reviews Microbiology. 2009;7:287–296. doi: 10.1038/nrmicro2122. [DOI] [PubMed] [Google Scholar]
- 13.Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Chen L, Fan X, Bian Y. De novo assembly of Auricularia polytricha transcriptome using Illumina sequencing for gene discovery and SSR marker identification. PLoS One. 2014;9:e91740. doi: 10.1371/journal.pone.0091740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu, Y. P. et al. De Novo Sequencing and Transcriptome Analysis of Pleurotus eryngii subsp. tuoliensis (Bailinggu) Mycelia in Response to Cold Stimulation. Molecules21, 10.3390/molecules21050560 (2016). [DOI] [PMC free article] [PubMed]
- 16.Trick M, Long Y, Meng J, Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol J. 2009;7:334–346. doi: 10.1111/j.1467-7652.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- 17.Schuster SC. Next-generation sequencing transforms today’s biology. Nat Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 18.Tang LH, et al. Transcriptome analysis of candidate genes and signaling pathways associated with light-induced brown film formation in Lentinula edodes. Appl Microbiol Biotechnol. 2013;97:4977–4989. doi: 10.1007/s00253-013-4832-y. [DOI] [PubMed] [Google Scholar]
- 19.Zhong M, et al. De novo characterization of Lentinula edodes C(91-3) transcriptome by deep Solexa sequencing. Biochem Biophys Res Commun. 2013;431:111–115. doi: 10.1016/j.bbrc.2012.12.065. [DOI] [PubMed] [Google Scholar]
- 20.Yu GJ, et al. Deep insight into the Ganoderma lucidum by comprehensive analysis of its transcriptome. PLoS One. 2012;7:e44031. doi: 10.1371/journal.pone.0044031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, et al. Transcriptome and proteome exploration to provide a resource for the study of Agrocybe aegerita. PLoS One. 2013;8:e56686. doi: 10.1371/journal.pone.0056686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin Y, et al. Genome-wide transcriptome and proteome analysis on different developmental stages of Cordyceps militaris. PLoS One. 2012;7:e51853. doi: 10.1371/journal.pone.0051853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pertea G, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 25.Robinson MD, McCarthy DJ, Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liers C, Bobeth C, Pecyna M, Ullrich R, Hofrichter M. DyP-like peroxidases of the jelly fungus Auricularia auricula-judae oxidize nonphenolic lignin model compounds and high-redox potential dyes. Appl Microbiol Biotechnol. 2010;85:1869–1879. doi: 10.1007/s00253-009-2173-7. [DOI] [PubMed] [Google Scholar]
- 27.Floudas D, et al. The Paleozoic Origin of Enzymatic Lignin Decomposition Reconstructed from 31 Fungal Genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 28.Yao FJ, et al. Development of a Molecular Marker for Fruiting Body Pattern in Auricularia auricula-judae. Mycobiology. 2018;46:72–78. doi: 10.1080/12298093.2018.1454004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu LX, et al. Construction of a genetic linkage map and QTL mapping of agronomic traits in Auricularia auricula-judae. J Microbiol. 2017;55:792–799. doi: 10.1007/s12275-017-7241-6. [DOI] [PubMed] [Google Scholar]
- 30.Li H, et al. The Genome Sequences of 90 Mushrooms. Sci Rep. 2018;8:9982. doi: 10.1038/s41598-018-28303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan, Y., Wu, F., Si, J., Zhao, Y. F. & Dai, Y. C. Whole genome sequence of Auricularia heimuer (Basidiomycota, Fungi), the third most important cultivated mushroom worldwide. Genomics, 10.1016/j.ygeno.2017.12.013 (2017). [DOI] [PubMed]
- 32.Vallee M, et al. Physical, Predictive Glycaemic Response and Antioxidative Properties of Black Ear Mushroom (Auricularia auricula) Extrudates. Plant Foods Hum Nutr. 2017;72:301–307. doi: 10.1007/s11130-017-0621-6. [DOI] [PubMed] [Google Scholar]
- 33.Meng X, Liang H, Luo L. Antitumor polysaccharides from mushrooms: a review on the structural characteristics, antitumor mechanisms and immunomodulating activities. Carbohydr Res. 2016;424:30–41. doi: 10.1016/j.carres.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Chen JZ, Zhang XD, Jiang ZY. The Application of Fungal Beta-glucans for the Treatment of Colon Cancer. Anti-Cancer Agents in Medicinal Chemistry. 2013;13:725–730. doi: 10.2174/1871520611313050007. [DOI] [PubMed] [Google Scholar]
- 35.Deng C, et al. Chemical analysis and antioxidant activity in vitro of a beta-D-glucan isolated from Dictyophora indusiata. Int J Biol Macromol. 2012;51:70–75. doi: 10.1016/j.ijbiomac.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Ren L, Perera C, Hemar Y. Antitumor activity of mushroom polysaccharides: a review. Food & Function. 2012;3:1118–1130. doi: 10.1039/c2fo10279j. [DOI] [PubMed] [Google Scholar]
- 37.Ma ZC, Wang JG, Zhang L, Zhang YF, Kan D. Evaluation of water soluble β-D-glucan from Auricularia auricular-judae as potential anti-tumor agent. Carbohydrate Polymers. 2010;80:977–983. doi: 10.1016/j.carbpol.2010.01.015. [DOI] [Google Scholar]
- 38.OuYang F, et al. AKT signalling and mitochondrial pathways are involved in mushroom polysaccharide-induced apoptosis and G1 or S phase arrest in human hepatoma cells. Food Chem. 2013;138:2130–2139. doi: 10.1016/j.foodchem.2012.10.047. [DOI] [PubMed] [Google Scholar]
- 39.Panthong, S., Boonsathorn, N. & Chuchawankul, S. Antioxidant activity, anti-proliferative activity, and amino acid profiles of ethanolic extracts of edible mushrooms. Genet Mol Res15, 10.4238/gmr15048886 (2016). [DOI] [PubMed]
- 40.Zhou Q, et al. Comparative analysis between low- and high-cadmium-accumulating cultivars of Brassica parachinensis to identify difference of cadmium-induced microRNA and their targets. Plant and Soil. 2017;420:223–237. doi: 10.1007/s11104-017-3380-0. [DOI] [Google Scholar]
- 41.Min XJ, et al. Transcriptome differences between enrofloxacin-resistant and enrofloxacin-susceptible strains of Aeromonas hydrophila. Plos One. 2017;12:e0179549. doi: 10.1371/journal.pone.0179549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao FX, et al. Distinct herpesvirus resistances and immune responses of three gynogenetic clones of gibel carp revealed by comprehensive transcriptomes. BMC Genomics. 2017;18:561. doi: 10.1186/s12864-017-3945-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, et al. Involvement of an ABI-like protein and a Ca2+ -ATPase in drought tolerance as revealed by transcript profiling of a sweetpotato somatic hybrid and its parents Ipomoea batatas (L.) Lam. and I. triloba L. PLoS One. 2018;13:e0193193. doi: 10.1371/journal.pone.0193193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang H, et al. Candidate odorant binding proteins and chemosensory proteins in the larval chemosensory tissues of two closely related noctuidae moths, Helicoverpa armigera and H. assulta. PLoS One. 2017;12:e0179243. doi: 10.1371/journal.pone.0179243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozawa R, et al. Intraspecific variation among Tetranychid mites for ability to detoxify and to induce plant defenses. Sci Rep. 2017;7:43200. doi: 10.1038/srep43200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colinet D, et al. Extensive inter- and intraspecific venom variation in closely related parasites targeting the same host: the case of Leptopilina parasitoids of Drosophila. Insect Biochem Mol Biol. 2013;43:601–611. doi: 10.1016/j.ibmb.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Mathe-Hubert H, et al. Comparative venomics of Psyttalia lounsburyi and P. concolor, two olive fruit fly parasitoids: a hypothetical role for a GH1 beta-glucosidase. Sci Rep. 2016;6:35873. doi: 10.1038/srep35873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie C, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan X, Zhou Y, Xiao Y, Bian Y. Cloning and characterization of two allelic glyceraldehyde-3-phosphate dehydrogenase genes in Auricularia auricula-judae. World journal of microbiology & biotechnology. 2014;30:181–189. doi: 10.1007/s11274-013-1436-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.