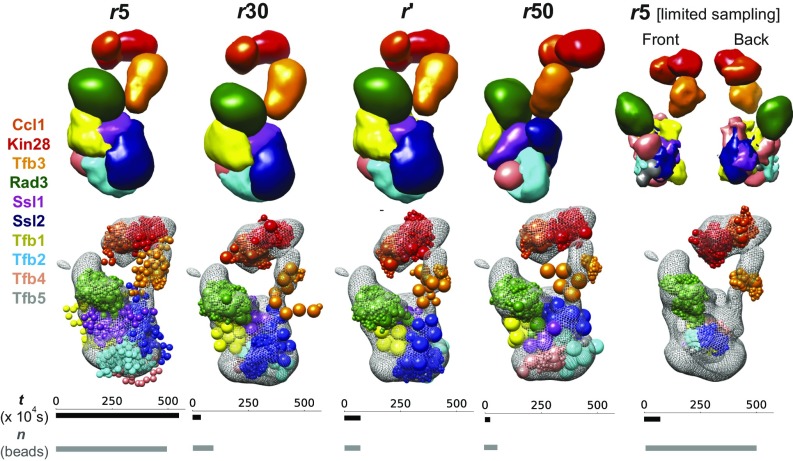
Fig. 3.
Comparison of representations of a large assembly. Comparing the performance of an approximately optimal representation (r′) with other uniform-resolution representations of 5 (r5), 30 (r30), and 50 (r50) residues per bead for the 10-protein transcription initiation and DNA repair factor TFIIH. Also shown is the performance of a five-residue per bead representation with the sampling time equal to that used for computing r′ and sampling the corresponding models (r5 [limited sampling]). Localization probability density maps specifying the probability of any volume element being occupied by a given bead in superposed good-scoring models (Top), EM map (Bottom, gray mesh), and representative models (Bottom, beads colored by protein) from the most populated cluster are shown for various representations. The total CPU time in seconds (t) for sampling models in various representations is shown as black bars (six-core dual Intel Xeon E5-2620 v3 processor). The total number of beads (n) for regions of unknown structure in each representation is shown in gray bars.