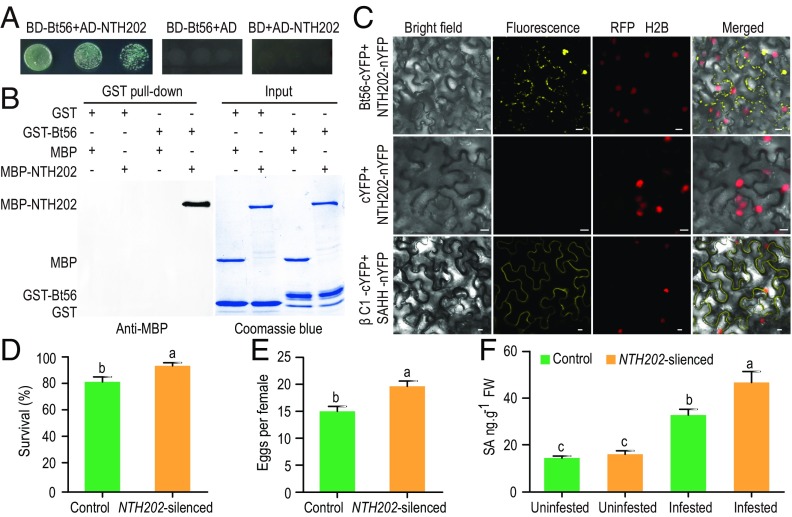
Fig. 4.
Interaction between Bt56 and plant NTH202 proteins. (A) Interaction between Bt56 and NTH202 in the Y2H. Yeast strain Y2H gold cotransformed with the indicated plasmids was spotted on the quadruple dropout media. The EVs pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. (B) GST pull-down assays. Glutathione S-transferase (GST) or GST-Bt56 fusion proteins were used to pull down maltose-binding protein (MBP) or MBP-NTH202 fusion proteins. Western blot was performed using an anti-MBP antibody to detect the associated proteins. Sodium dodecyl sulfate polyacrylamide gel electrophoresis gel was stained with CB to monitor the input protein amount. (C) BiFC analysis of Bt56 interaction with NTH202 in vivo. Fluorescence was observed as complementation of the Bt56-cYFP fusion protein and the NTH202-nYFP fusion protein. Red fluorescent protein- (RFP-) histone 2B (RFP-H2B) was used as a marker for the nucleus. Unfused cYFP was coexpressed with NTH202-nYFP as a negative control, and βC1 was expressed with S-adenosyl homocysteine hydrolase (SAHH) as a positive control (55). The BiFC assays were repeated three times, and a total of 24 images were analyzed. Bars = 10 μm. The survival (D) and fecundity (E) of whiteflies feeding on the control and NTH202-silenced tobacco. Each treatment included nine plants, and each plant had two clip cages (n = 18). (F) Mean level of SA in the control, NTH202-silenced, whitefly-infested, and NTH202-silenced plus whitefly-infested tobacco plants (n = 7 to 8). Fresh weight. Values shown are means ± SE. The letters above the bars indicate significant differences among different treatments at P < 0.05 (nested ANOVA for whitefly bioassays; one-way ANOVA followed by the LSD test for other experiments).