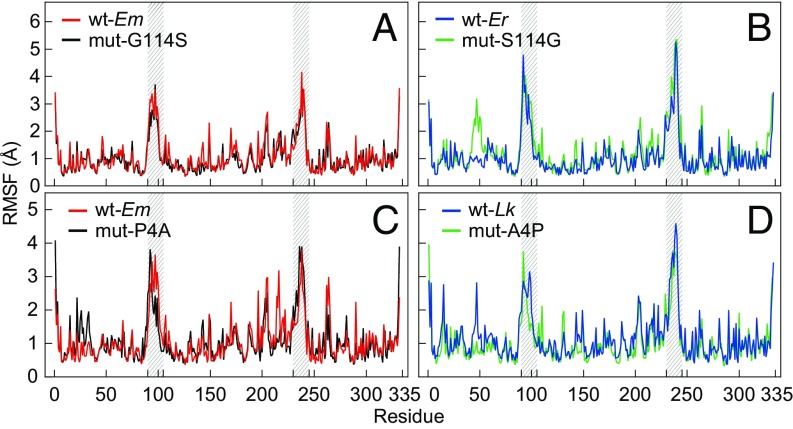
Fig. 7.
The rms fluctuation (rmsf) for individual residues over the equilibration state (10–20 ns) of cMDHs at a simulation temperature of 57 °C. (A and B) Two wild-type enzymes, Echinolittorina malaccana (wt-Em) and E. radiata (wt-Er), and two mutant enzymes, mut-G114S (glycine mutated to serine at site 114 for the E. malaccana ortholog) and mut-S114G (serine mutated to glycine for the E. radiata ortholog) (n = 10). (C and D) Two wild-type enzymes, wt-Em and Littorina keenae (wt-Lk), and two mutant enzymes, mut-P4A (proline mutated to alanine at site 4 for the E. malaccana ortholog) and mut-A4P (alanine mutated to proline for the L. keenae ortholog) (n = 5). The locations of the two mobile regions (residues 90–105 for MR1 and residues 230–245 for MR2) are highlighted by gray shading.