Fig. 5.
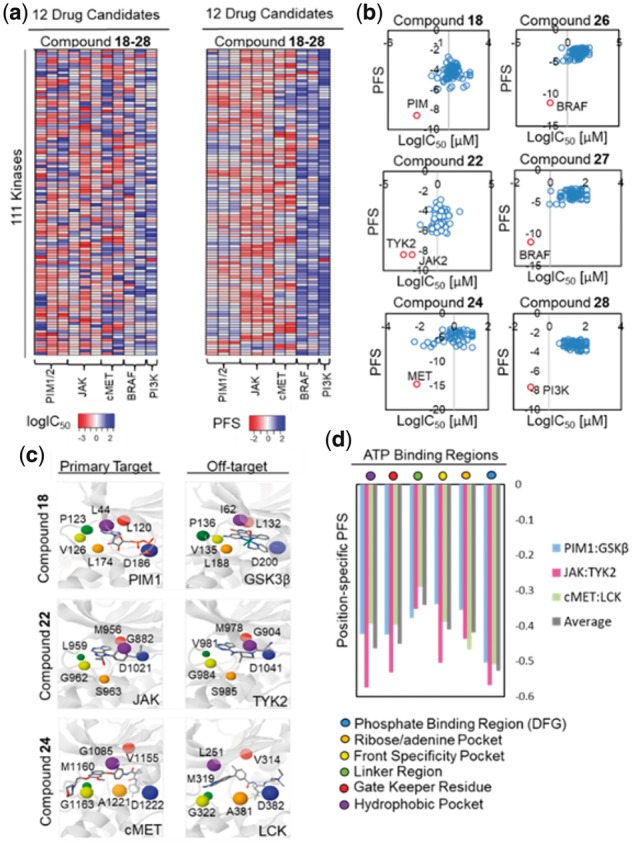
Off-target identification of kinase drug candidates. (a) Heatmaps of kinase off-target binding profile generated based on PFS values or experimentally observed IC50 values (logIC50) for the 11 kinase drug candidates (compounds 18–28). Consistent with the experimental data, predicted PFS revealed promiscuous kinase off-target binding for PIM inhibitors (compounds 18, 19 and 20), JAK inhibitors (compounds 21, 22 and 23), and cMET inhibitors (compounds 24 and 25) while binding for BRAF inhibitors (compounds 26 and 27), and a PI3K inhibitor (compound 28) were highly specific. (b) Scatterplots of predicted PFS values and experimental IC50 values (logIC50) for PIM inhibitor (compound 18), JAK inhibitor (compound 22), cMET inhibitor (compound 24), BRAF inhibitor (compound 26 and 27), and PI3K inhibitor (compound 28). Note that the primary targets with the highest binding affinity (lowest logIC50 values) were predicted by the optimal (most negative) PFS. (c) Structural alignments between primary targets: PIM1 (PDB: 1XR1), JAK (PDB: 4IVD), and cMET (PDB: 3LQ8) and off-targets: GSK3β (PDB: 3PUP), TYK2 (PDB: 3LXN), and LCK (PDB: 2OG8) with CHK1 kinase (PDB: 1NVR) identified critical residues pair involved in kinase off-target binding from the multiple-sequence alignment (see Supplementary Fig. S12). (d) The identified critical binding regions include the phosphate binding region/DFG, the ribose/adenine pocket, the front specificity pocket, the linker region, the gate keeper residue, and the hydrophobic pocket region. Site similarity comparison based on the PocketFEATURE algorithm showed that phosphate binding region/DFG, hydrophobic pocket, and gate keeper regions have the highest average PS-PFS, suggesting their important functional roles in modulating compound cross-activities (Fig. 5d and Supplementary Fig. S12)
