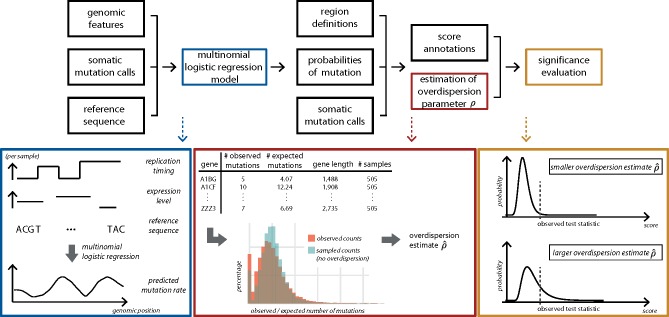
Fig. 1.
Overview of ncdDetect v.2. For each sample, the genomic features replication timing, expression level and reference sequence are used as explanatory variables to predict the sample- and position specific probabilities of mutation in a multinomial logistic regression model (blue box). For a specific genomic region type, the observed and expected number of mutations are collected for each specific candidate element (red box; illustrated for protein-coding genes). This information is applied to estimate the overdispersion parameter ρ as explained in Section 2.4. The estimated amount of overdispersion is accounted for in the significance evaluation of each candidate element, as explained in Section 2.2. The larger the overdispersion estimate, the harder it will be for a genomic candidate element to reach significance (yellow box)