Fig. 3.
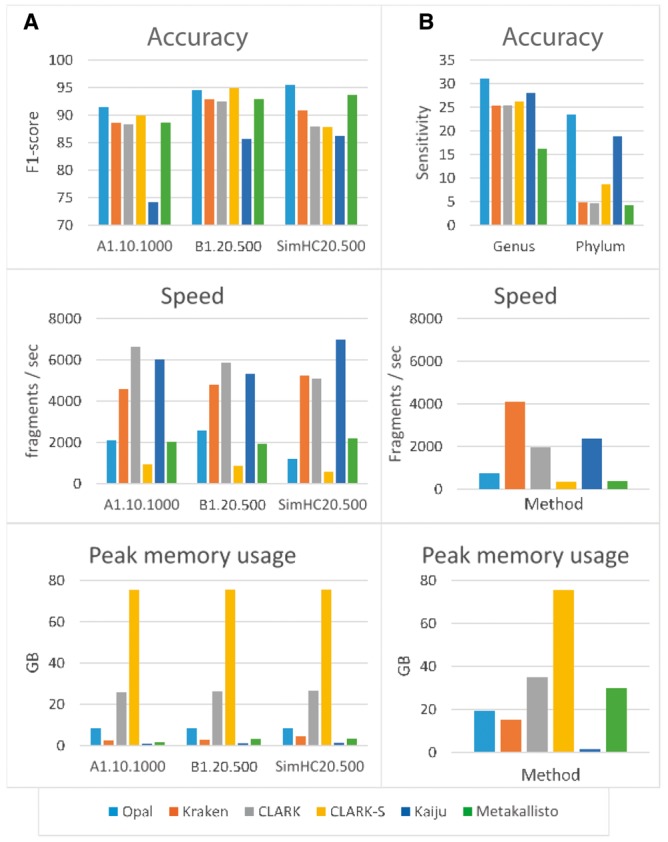
Comparison of Opal against Kraken, CLARK, CLARK-S, Kaiju and Metakallisto in terms of accuracy (top row), speed (middle row), and memory usage (last row): (a) Opal achieves generally higher classification accuracies on three public benchmark data sets than five other state-of-the-art compositional classifiers. Only CLARK-S is comparable in terms of accuracy, but CLARK-S uses an order of magnitude more memory while running significantly slower (processes fewer fragments/sec). (b) Opal has greater sensitivity to novel lineages in benchmarks on a large 193-species dataset [19]. We simulate the effect of novel species by removing a species from the dataset, and training at the genus level on the remaining data. Then, we predict the genus of simulated reads from the removed species. Similarly, we repeated the experiment removing all data from a genus, training at the phylum level, and attempting to predict the phylum of the removed genus. The improvement over Kaiju is particularly impressive as Kaiju bins using protein sequences, giving it an inherent advantage at higher phylogenetic levels, which we overcome using low-density hashing
