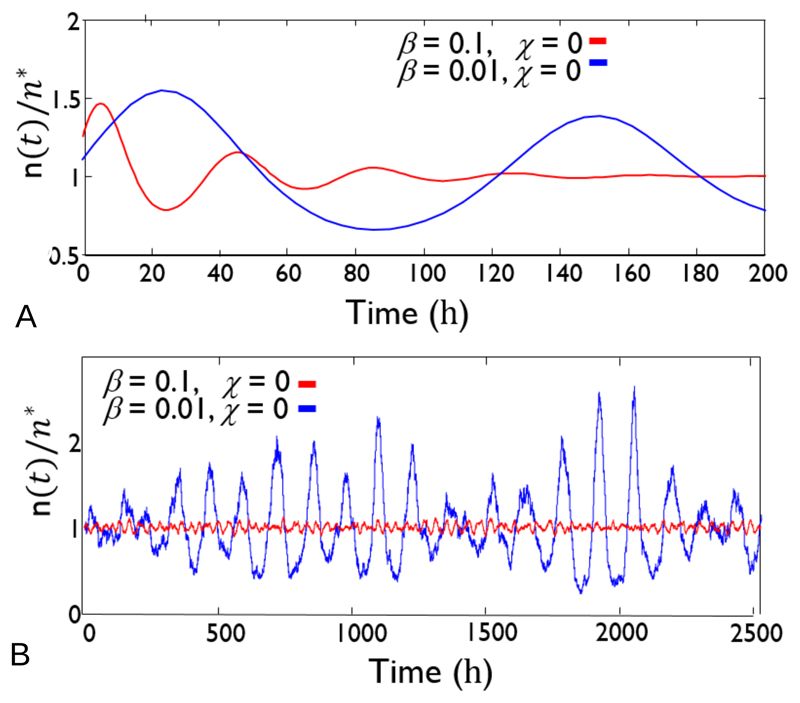
Figure 4.
Dynamical predictions for the population density of E. coli bacteria growing on glucose, with parameters rmax = 1hr−1, d = 0.5hr−1, Ks = 1μM and Γ = 1.8 × 1010 glucose molecules per bacterium. The nutrient inflow rate b is varied, such that we have β = 0.1, χ = 0 (corresponding to b = 0.1μMh−1 and R=0; such that s* = 1μM and n* ≈ 107 bacteria per litre) and β = 0.01, χ = 0 (corresponding to b = 0.01μMh−1 and R=0; such that s* = 1μM and n* ≈ 106 bacteria per litre). Panel A shows results for the deterministic model, Eqs. (18-19). Panel B shows results for the stochastic model, Eq. (22), for a system volume of 1ml; i.e. for approximate absolute bacterial numbers of 104 (red) and 103 (blue). Note the different time axes in the two panels. In these simulations, the bacterial densities are much lower than in a typical microbiology lab experiment, and are 1-2 orders of magnitude lower than the bacterial density in seawater, but they are similar to the bacterial density that might be found in drinking water.