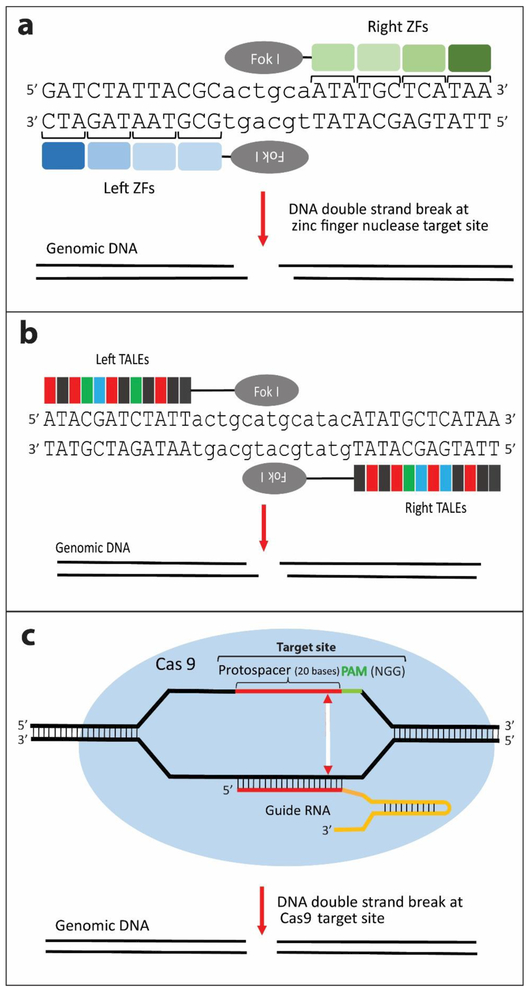
Fig. 1. Programmable nucleases for site-specific cleavage in the genome.
(a) ZFNs: Each zinc finger (ZF) binds to 3 nucleotides of DNA. An array of 3-4 ZFs is coupled with the FokI nuclease for cleavage. (b) TALENs: Each TALE domain binds to a single nucleotide. An array of TALEs is coupled with the FokI nuclease. For both (a) and (b), FokI nuclease acts as an obligate heterodimer where the nuclease domain cleaves the spacer sequence to create a DSB with cohesive ends, (c) The sgRNA contains a 19–20 base sequence (red) within the CRISPR RNA (crRNA) that is complementary to the chromosomal target site, and this is combined with a transactivating CRISPR RNA (tracrRNA) (yellow) to guide the Cas9 nuclease (blue oval) to the target site where it creates DSBs with blunt ends 3 bases upstream of the PAM sequence (green).