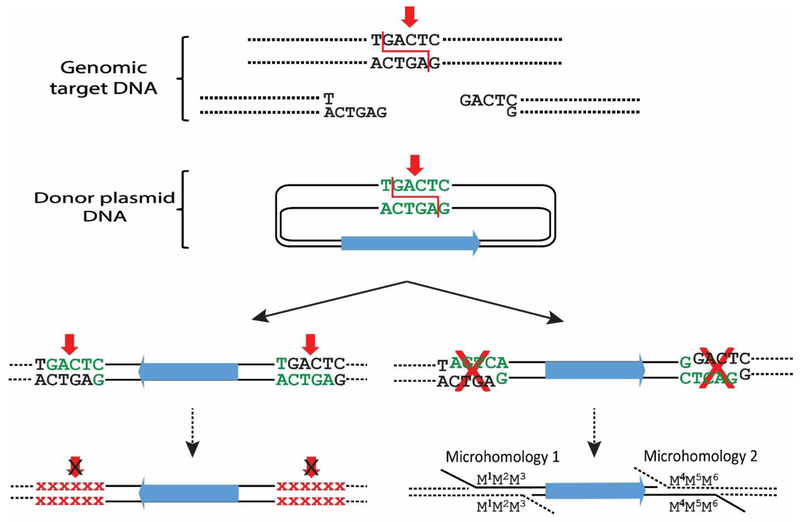
Fig. 3. End-capture of cohesive ends.
Diagram of “end capture” of a piece of donor DNA at the genomic site of a nuclease-induced (red arrows) DSB. Two pathways could result in stable products, but at low efficiency and with imprecise junctions. Left: The overhanging ends of the donor DNA are complementary to the overhanging ends of the genomic target site, since both the donor and genomic DNAs were cleaved by the same ZFN. However, end capture recreates the target sites that can be re-cleaved. Recutting and religation will be repeated until the target site sequence is altered by mutation and can no longer be cleaved by the programmable nuclease. Small red Xs indicate the mutated sequences. Right: Donor DNA that inserts with an opposite orientation to that shown in the left pathway will have incompatible ends to the genomic target sites, but it can be integrated via microhomology with very low efficiency.