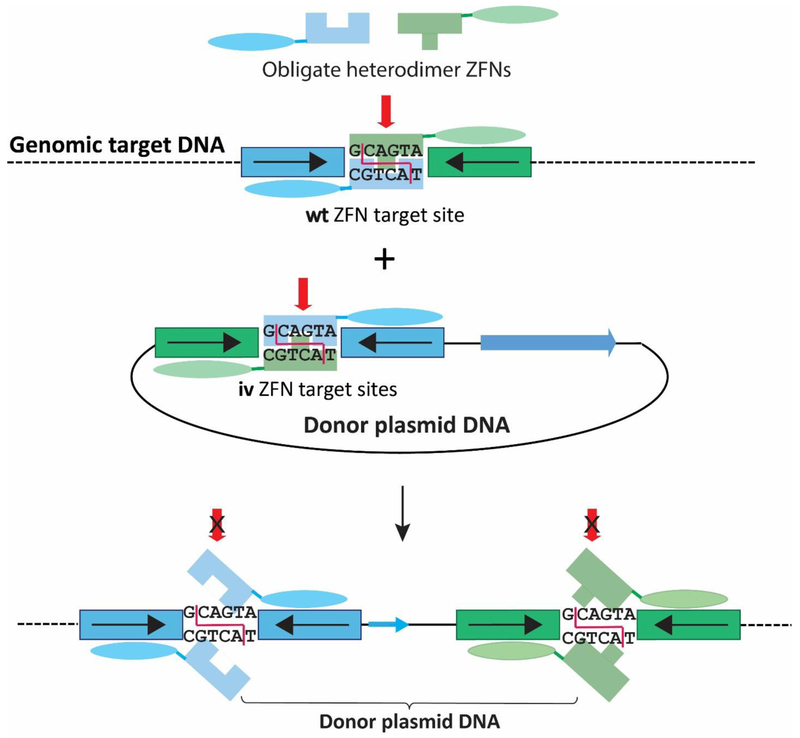
Fig. 5. Obligate ligation-gated recombination (ObLiGaRe).
A pair of ZFNs cleaves both the wild type (wt) target site of genomic DNA and the inverted target sites (iv) of the donor construct. Both the wt and iv target sites have the identical spacer sequence between the left and right ZFN binding sites, thus creating complementary overhanging ends between the donor DNA and genomic target for end-capture. Once the linearized donor has been ligated into the genomic target site, the same pair of ZFNs cannot cut (x over red arrow) the newly formed junctions since homodimer formation is prevented by the use of an obligatory heterodimeric FokI nuclease domain. Moreover, the inverted target sequence in the donor plasmid prevents adjacent placement of the green and blue sequences. Thus, the efficiency of formation of the final product of the integrated donor DNA at the genomic target site is increased as re-cutting cannot occur. The single dotted or solid lines represent dsDNA of genomic or plasmid DNA, respectively. In the example shown here, the entire donor plasmid is integrated into the target site.