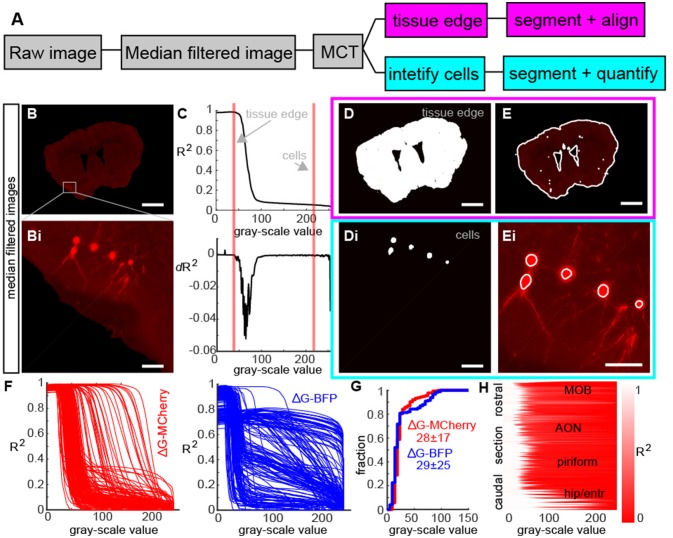
Figure 2.
Image processing steps for whole brain reconstructions. (A) Flow diagram of image processing for identifying tissue structure and neurons. (B) Representative example of a median filtered fluorescence image section and (Bi) enlargement of retrogradely labeled neurons showing blurring of cell bodies due to filtering. (C, top) Correlation of a thresholded image to the median filtered image as a function of threshold value and (C, bottom) derivate of correlation curve. Red lines show transitions in the image statistics that correspond to the tissue edge which identifies the section boundaries and labeled cells. (D) Example of thresholded images from A at the values corresponding to the tissue edge and (Di) to retrogradely labeled neurons. (E) Outline of tissue edge and (Ei) individual cells shows that the algorithm identifies both the sections in the imaging and individual neurons. (F) Correlation curve (as described in C) for all sections in an experiment using G-MCherry (left) and G-BFP show the diversity of threshold functions across different sections and different viral constructs. (G) Cumulative histogram of threshold values selected for each section in (F) for G-MCherry and G-BFP highlight this diversity. (H) Correlation curves as a function of rostral-caudal axis shows a systematic variation from the olfactory bulb to the hippocampus and entorhinal cortex, reflecting the ability of the algorithm to segment sections and identify cells across the systematic differences in fluorescence associated with different sections.