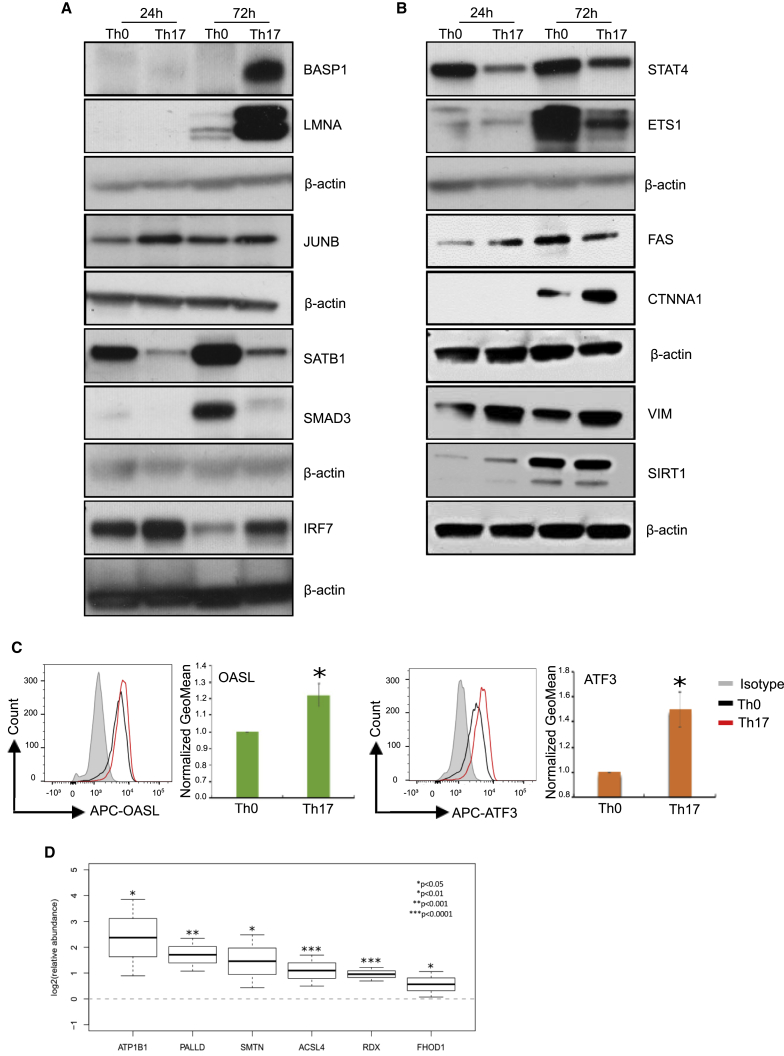
Figure 4.
Validation of the Mass-Spectrometry (MS)-Identified Proteins Using Immunoblotting, Flow Cytometry, and Targeted Mass-Spectrometry-Based Selected Reaction Monitoring (SRM) Analysis
(A and B) Immunoblot validations of MS identified up- or downregulated proteins with known and unknown Th17-related function. Blots show the protein extracts from the Th0 and Th17 cells at 24 and 72 hr. A representative blot from three biological replicates is shown.
(C) Flow cytometry validations of OASL and ATF3 detected as differentially upregulated in Th17 cells by MS analysis. A representative histogram image from Th17 (red), Th0 (black), and isotype control (gray) as well as the mean of four biological replicates are shown. Error bars represent standard deviation values across biological replicates (n=4; *p<0.05 for Th0 versus Th17; two-tailed t-test analysis).
(D) Confirmation of the protein expression of six proteomics candidates, ATP1B1, PALLD, ACSL4, FHOD1, SMTN, and RDX, using targeted SRM analysis. The SRM data is from four biological replicates and represented as the mean values +/- 95% confidence interval. (n = 4; *p < 0.05, **p < 0.01, ***p < 0.0001, for Th17 versus Th0).