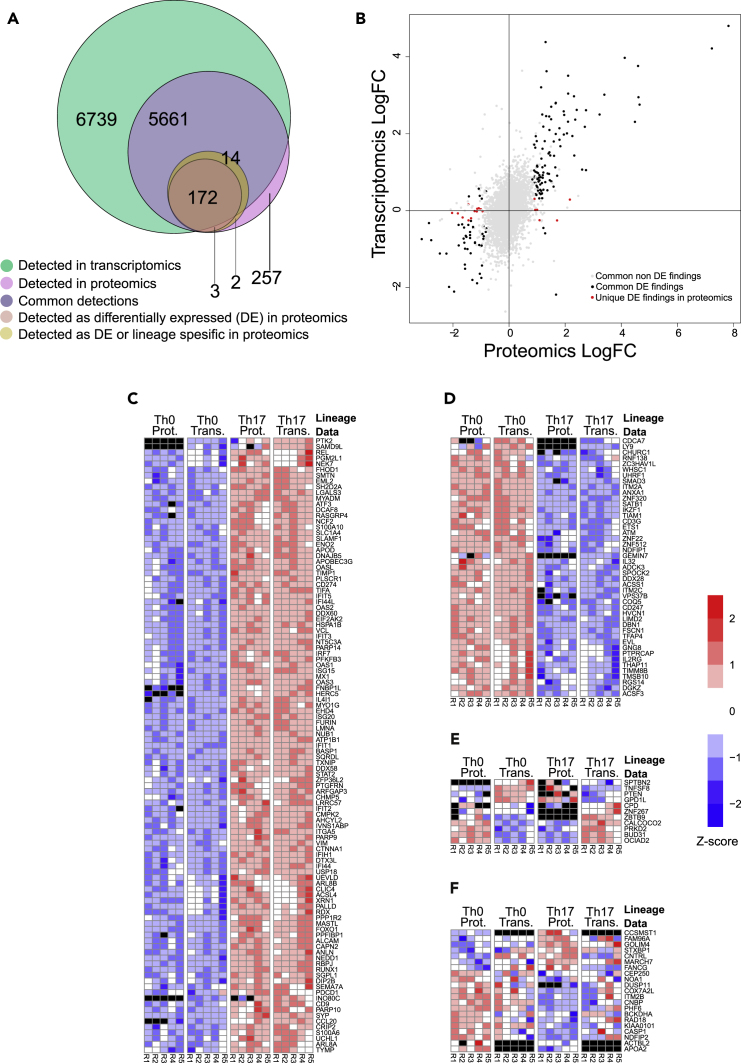
Figure 5.
Correspondence of the Differentially Regulated Proteins between Th17 and Th0 and the Transcriptomics Data at 72 hr
(A) Venn diagram demonstrating the total number of common and unique detections and the total number of differentially regulated and differentially expressed (DE) proteins detected uniquely in the proteomics and commonly with the transcriptomics data. Differentially regulated proteins refer to those proteins detected as differentially expressed or detected only in one condition.
(B–E) (B) The logarithmic fold changes (logFC) of the all the common detections in proteomics and transcriptomics. The differentially expressed (DE) proteins detected as DE also in transcriptomics are marked with black and those uniquely DE in proteomics are marked with red. The Pearson correlation coefficient over the logFCs over all the common detections was 0.519 (p value < 0.001, n = 5,661), over the common DE findings 0.825 (p<0.001, n = 154), and over the DE findings unique to proteomics among the common detections 0.233 (p = 0.35, n = 18). The Z score standardized expression levels of the DE proteins and the corresponding DE transcripts between Th17 and Th0 (C) upregulated in Th17 in both datasets, (D) downregulated in Th17 in both datasets, and (E) divergently regulated between datasets.
(F) The Z score standardized expression levels of the DE proteins not detected as DE in transcriptomics. Black color in the heatmaps stands for undetected/missing value.