Figure 6.
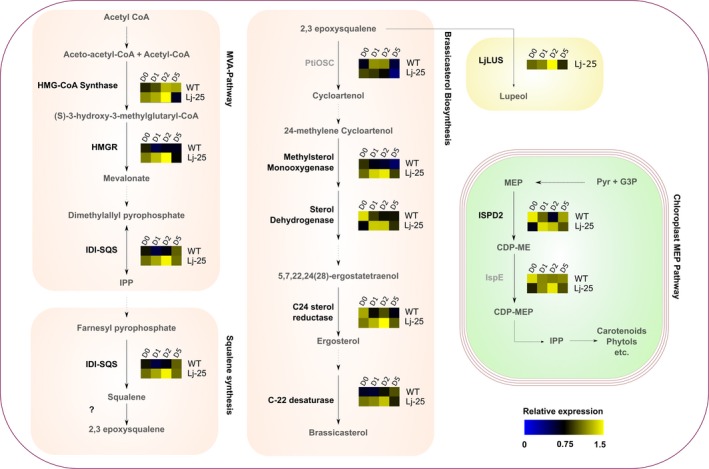
mRNA expression of sterol biosynthesis enzymes during a five‐day time course in the wild type (WT) and the LjLUS‐25 line. Schematic representation of sterol metabolism in Phaeodactylum tricornutum, indicating the gene selected for mRNA expression analysis and relative heat map for WT and LjLUS‐25 line. Values are the expression relative to the geometric average of the RP3a and UBQ reference genes. Transcripts not significantly different between wt and LjLUS are indicated in grey. Enzymes involved in the cytosolic MVA pathway: HMGS: hydroxymethylglutaryl‐CoA synthase; HMGR: 3‐hydroxy‐3‐methyl‐glutaryl‐coenzyme A reductase; IDI‐SQS: isopentenyl diphosphate isomerase‐squalene synthase. Enzymes involved in brassicasterol biosynthesis: PtOSC: oxidosqualene cyclase, methylsterol monooxygenase, sterol dehydrogenase, C24 sterol reductase, C22 sterol desaturase. Enzymes involved in the plastidial MEP pathway: ISPD2: 2‐C‐methyl‐d‐erythritol 4‐phosphate cytidylyltransferase, ISP‐E: 4‐diphosphocytidyl‐2‐C‐methyl‐d‐erythritol kinase. In the yellow box, the non‐native lupeol synthase from Lotus japonicus, introduced in our engineered strain LjLUS25. Dashed lines indicate multiple reactions. Gene accession numbers are mentioned in the results section. Precursor abbreviations: IPP: Isopentyl pyrophosphate, MEP: 2‐C‐methyl‐d‐erythritol 4‐phosphate, CDP‐ME: 4‐diphosphocytidyl‐2‐c‐methylerythritol, CDP‐MEP: 4‐diphosphocytidyl‐2‐c‐methyl‐d‐erythritol 2‐phosphate, DMAP: dimethylallyl pyrophosphate, G3P: glycerol 3‐phosphate, Pyr: pyruvate.
