Extended Data Figure 8. Tetsting SLC16A1 in efferocytosis.
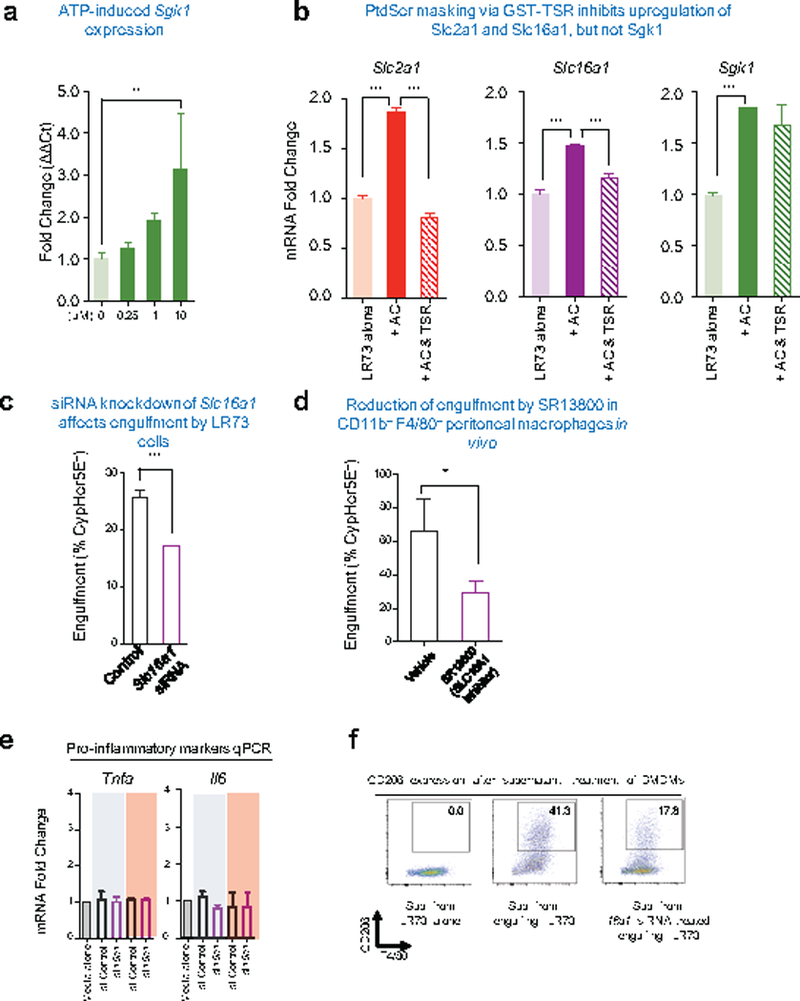
(a) qPCR determination of Sgk1 expression phagocytes treated with ATP. LR73 cells were treated with indicated amount of ATP for 4h. Expression of Sgk1 was determined by qPCR using hamster-specific primers. **p < .01. Data are representative of at least two independent experiments with 3–4 replicates per condition.
(b) qPCR determination of Slc2a1, Slc16a1, and Sgk1 expression in phagocytes after addition of the PtdSer masking peptide (GST-TSR) during efferocytosis. Apoptotic cells (AC) were added with or without TSR peptide (10ng/μl) for 4hr. Expressions of indicated genes were determined by qPCR using hamster-specific primers. ***p < .001. Data are representative of at least two independent experiments with 3–4 replicates per condition.
(c) SLC16A1 inhibition blocks efferocytosis in vitro. LR73 cells were treated with Slc16a1 siRNA and uptake of CypHer5E labeled apoptotic Jurkat cells assessed. ***p < .001.
(d) SLC16A1 inhibitor SR13800 dampens efferocytosis by peritoneal macrophages. C57BL/6 mice were intraperitoneally injected with SR13800 (10mg/kg) in X-VIVO media for 1h prior to apoptotic cell injection. CypHer5E-labeled apoptotic Jurkat cells were injected intraperitoneally along with the drug. After 1hr, apoptotic cell engulfment by CD11b+ F4/80hi peritoneal macrophages was analyzed by flow cytometry. *p < .05. Data are representative of two independent experiments with at least 6 mice in each group per experiment.
(e and f) Supernatants from control LR73 or Slc16a1 siRNA-treated LR73 cells engulfing apoptotic cells was prepared, and added to BMDMs, and incubated for 12 h. Expression of inflammatory markers are determined by qPCR. ***p < .001. After 24 h, with supernatant incubation, CD206 and F4/80 expressions were determined by flow cytometry in BMDMs. Data are representative of two independent experiments with 2–3 replicates per condition.
