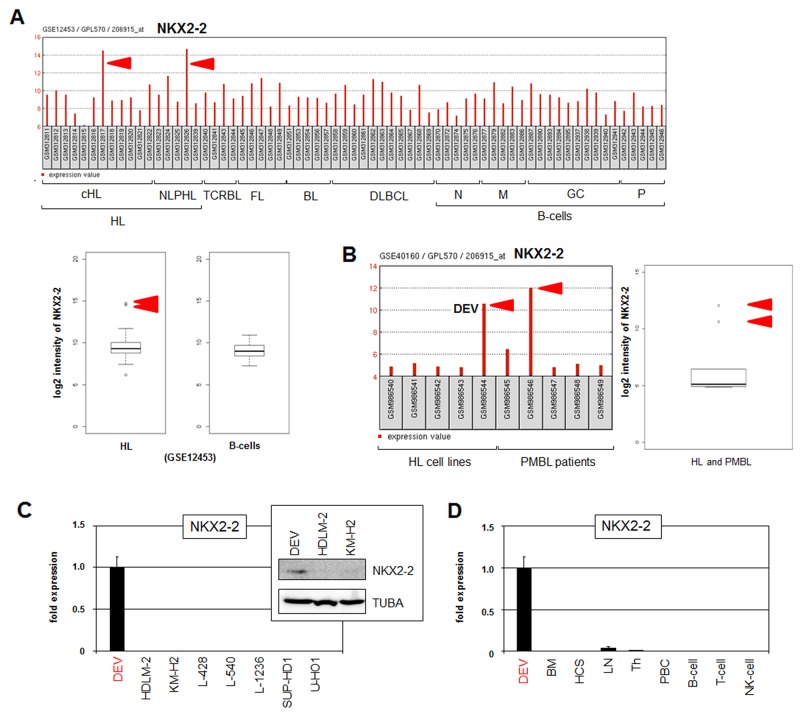
Figure 1. Expression of NKX2-2 in HL patients and cell lines.
(A) Expression profiling data of NKX2-2 (206915_at) in dataset GSE12453 containing samples of patients and normal B-cells are indicated as barplot (above) and boxplot (below). Hodgkin lymphoma (HL), T-cell rich B-cell lymphoma (TCRBL), follicular lymphoma (FL), Burkitt lymphoma (BL), diffuse large B-cell lymphoma (DLBCL), naïve B-cells (N), memory B-cells (M), germinal centre B-cells (GC), plasma cells (P). The red arrows indicate samples showing NKX2-2 overexpression detected in 1/12 (8%) cHL and in 1/5 (20%) NLPHL patients. (B) Expression profiling data of NKX2-2 in dataset GSE40160 containing samples of five HL cell lines including DEV and of five peripheral mediastinal B-cell lymphoma (PMBL) patients are indicated as barplot (left) and boxplot (right). The red arrows indicate DEV and one sample showing NKX2-2 overexpression, representing 20% of PMBL patients. (C) Expression analysis of NKX2-2 in HL cell lines by RQ-PCR and Western blot (insert). (D) RQ-PCR analysis of NKX2-2 in DEV in comparison to primary hematopoietic cell/tissue samples: bone marrow (BM), hematopoietic stem cell (HSC), lymph node (LN), thymus (Th), peripheral mononuclear blood cells (PBC), B-cells, T-cells and NK-cells.