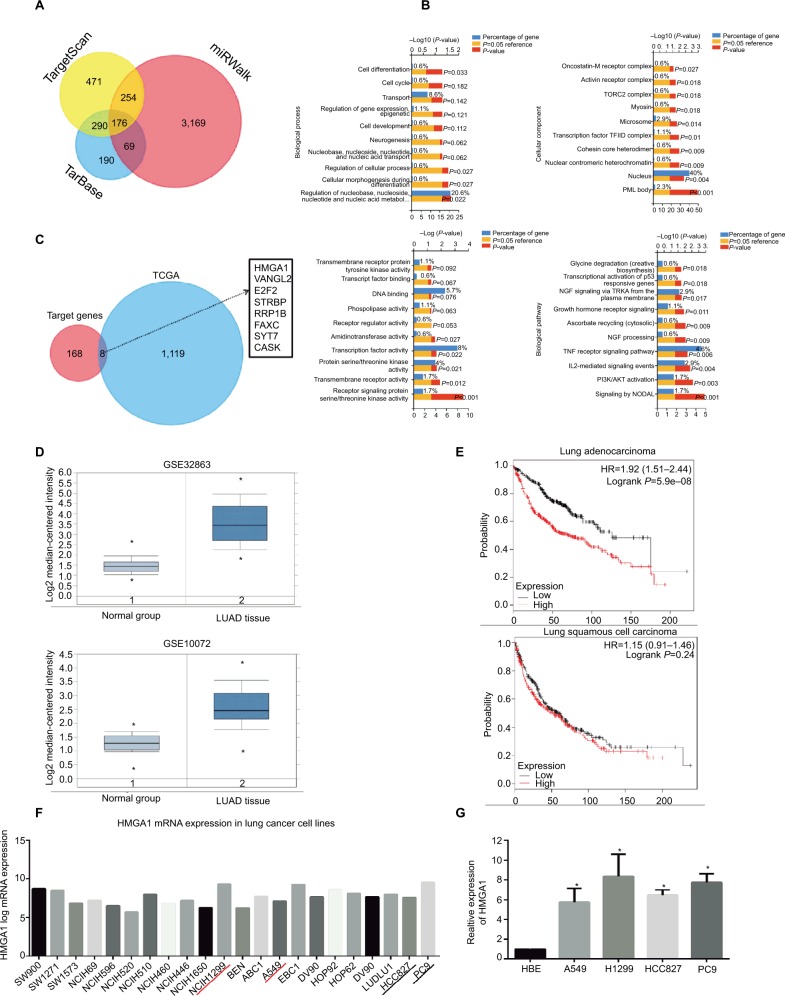
Figure 4.
Bioinformatics analysis of the targets of miR-4458.
Notes: (A) Venn chart of common target genes from three publicly available databases including TargetScan, TarBase, and miRwalk. (B) GO analysis included biological processes, cellular component, molecular function, and pathway enrichment analysis for 176 common targets. (C) Venn chart of 8 more possible targets from 176 common target genes and TCGA database. (D) The expression of HMGA1 between LUAD and normal group in GSE32863 and GSE10072 datasets. (E) Analysis of the OS about HMGA1 from Kaplan–Meier plotter database. (F) The expression of HMGA1 in lung cancer cell lines from CCLE database. (G) The expression of HMGA1 between four NSCLC cell lines (A549, H1299, HCC827, and PC9) and one normal lung cell (HBE) by qRT-PCR. *P<0.05, **P<0.01.
Abbreviations: GO, Gene Ontology; miR-4458, microRNA-4458; NC, negative control; OS, overall survival.