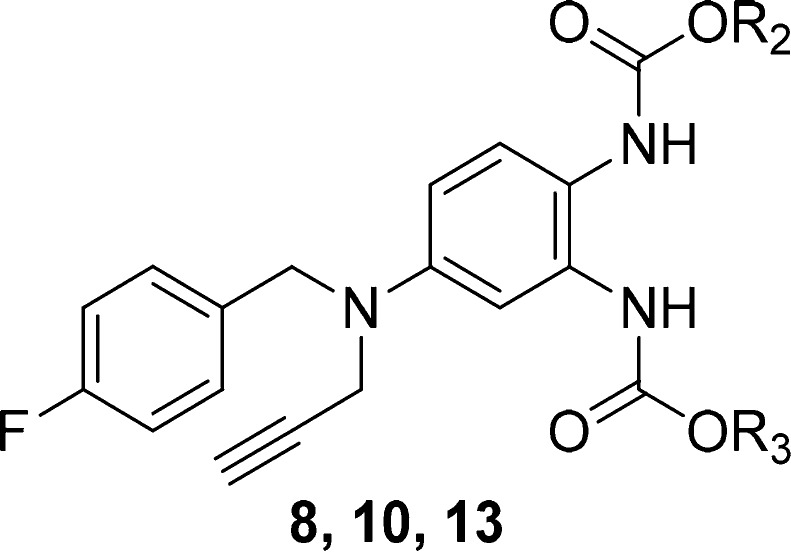
Table 2. Structures of N-2/3 Derivatives 8, 10, and 13 and Their Effects on the KCNQ2, 4, and 5 Channels.
|
I/I0a |
|||||
|---|---|---|---|---|---|
| Cpds | R2 | R3 | KCNQ2 | KCNQ4 | KCNQ5 |
| 8a | Me | Me | 1.37 ± 0.11 | 4.77 ± 0.44 | 2.89 ± 0.07 |
| 8b | allyl | allyl | 0.67 ± 0.02 | 2.21 ± 0.11 | 5.26 ± 0.53 |
| 8c | nPr | nPr | 0.78 ± 0.04 | 2.58 ± 0.56 | 2.75 ± 0.84 |
| 8d | iPr | iPr | 0.35 ± 0.04 | 1.33 ± 0.06 | 3.11 ± 0.93 |
| 8e | nBu | nBu | 0.77 ± 0.02 | 2.75 ± 0.40 | 4.13 ± 1.09 |
| 8f | iBu | iBu | 0.83 ± 0.04 | 1.52 ± 0.17 | 1.73 ± 0.10 |
| 8g | tBu | tBu | 1.00 ± 0.09 | 3.05 ± 0.74 | 5.32 ± 0.39 |
| 8h | cyclopropyl | cyclopropyl | 0.64 ± 0.03 | 1.51 ± 0.04 | 2.55 ± 0.38 |
| 8i | cyclobutyl | cyclobutyl | 0.26 ± 0.06 | 1.21 ± 0.09 | 1.33 ± 0.23 |
| 8j | cyclopentyl | cyclopentyl | 0.21 ± 0.05 | 0.88 ± 0.07 | 1.25 ± 0.14 |
| 8k | Ph | Ph | 0.91 ± 0.02 | 1.22 ± 0.05 | 1.16 ± 0.04 |
| 8l | Bn | Bn | 1.25 ± 0.01 | 3.02 ± 0.28 | 3.57 ± 0.31 |
| 10a | Et | Me | 0.61 ± 0.05 | 2.56 ± 0.47 | 3.34 ± 0.54 |
| 10b | Et | Allyl | 0.96 ± 0.04 | 3.68 ± 0.36 | 4.67 ± 0.69 |
| 10c | Et | nPr | 1.07 ± 0.03 | 3.95 ± 0.89 | 5.81 ± 0.62 |
| 10d | Et | iPr | 0.94 ± 0.06 | 5.01 ± 0.83 | 5.71 ± 0.34 |
| 10e | Et | nBu | 1.06 ± 0.03 | 4.16 ± 0.60 | 3.62 ± 0.31 |
| 10f | Et | iBu | 1.04 ± 0.04 | 5.19 ± 0.64 | 4.84 ± 0.32 |
| 10g | Et | tBu | 1.16 ± 0.04 | 6.37 ± 0.84 | 4.58 ± 0.32 |
| 10h | Et | Bn | 0.97 ± 0.15 | 1.27 ± 0.03 | 2.57 ± 0.67 |
| 13 | tBu | Et | 1.07 ± 0.06 | 7.71 ± 0.50 | 4.59 ± 0.56 |
The testing concentration was 10 μM. Each compound was tested in more than four cells.