Figure 1:
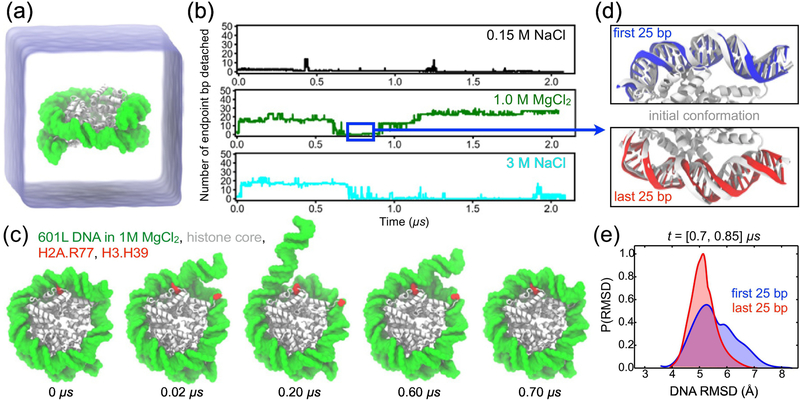
All-atom MD simulation of DNA breathing, (a) Simulation setup: a nucleosome particle composed of a histone core (white) and a 145-base pair (bp) fragment of DNA (green) is surrounded by electrolyte solution (semi-transparent blue), (b) The number of endpoint bp detached from the histone core as a function of time in the simulations of the Widom 601L nucleosome performed in 0.15 M NaCl (top), 1.0 M MgCl2 (middle), and 3.0 M NaCl (bottom). The number of endpoint bp detached was determined by finding all DNA-histone contacts, defining a contact as a pair of non-hydrogen atoms of the protein and DNA located within 4.5 A of each other, and identifying the contact-forming DNA bps closest to the entry and exit termini, the terminal contact bps. The total number of bp detached was calculated as the total number of bps in the nucleosomal DNA (i.e. 145) minus the number of bps located between the terminal contact bps. (c) Snapshots illustrating spontaneous unbinding and rebinding of nucleosomal DNA from and to the histone core observed in 1 M MgCl2 solution. Starting from a fully-bound state (0 μs), two DNA-histone contacts from the entry side break (0.02 μs, 0.20 μs) and later reform (0.60 μs, 0.70 μs). H2A Arg77 and H3 His39 are highlighted in red. (d) Reversibility of DNA detachment. Top and bottom panels illustrate the microscopic configurations of the two terminal fragments of DNA (first and last 25 bp, respectively) at the beginning (white) and after 0.8 μs (red and blue) of the MD simulation performed in 1.0 M MgCl2. The DNA fragment featured in the top panel unbinds from and rebinds to the histone core within the first 0.7 μs of the simulation, (e) Normalized distributions of the DNA fragments’ RMSD values for the [0.70, 0.85] μs segment of the 1.0 M MgCl2 trajectory. The RMSD values were computed with respect to the starting conformations using coordinates of all non-hydrogen DNA atoms, after alignment of the histone core backbone atoms.