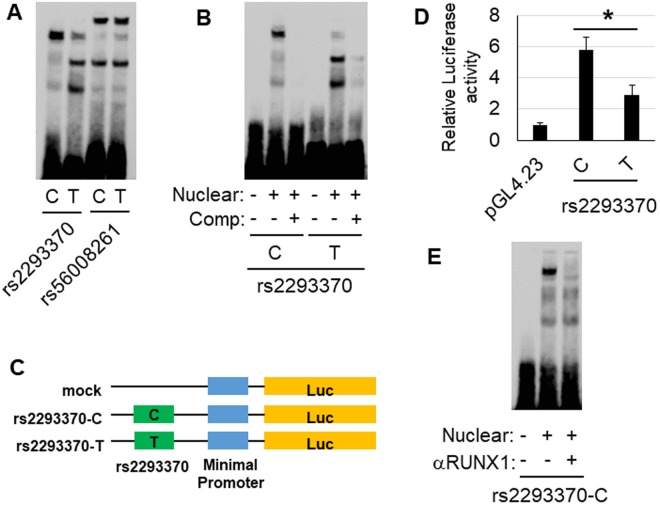
Figure 3.
In vitro functional analysis of each candidate variation in chromosome 3q13.33. (A) EMSA of each candidate primary variation using biotin-labeled probes corresponding to the major and the minor allele, and nuclear extracts of HepG2 cells. rs2293370 was the only SNP to show a difference in mobility shift between the two alleles. (B) Competitor assay, using HepG2 nuclear extracts and a 200× concentration of unlabeled probe corresponding to either the C (i.e., PBC susceptibility) or T alleles of rs2293370. (C) Outline of reporter plasmid constructs. PCR fragments of intron 2 of TIMMDC1, containing rs2293370, were sub-cloned into the pGL4.23 vector. (D) Transcription was measured by cellular luciferase activity, 24 h after transfection of HepG2 cells. The luciferase activities of cells transfected with the PBC susceptibility allele (C allele) of rs2293370 were higher than those transfected with the T allele. Three independent experiments with triplicate measurements were performed for each assay, and data represent mean ± SD; *P < 0.05 (Student’s t-test). (E) Identification of transcription factors targeting the C allele of rs2293370. A super-shift was observed following incubation of HepG2 cell nuclear extracts with an anti-RUNX1 antibody. Three independent experiments were performed in each assay.