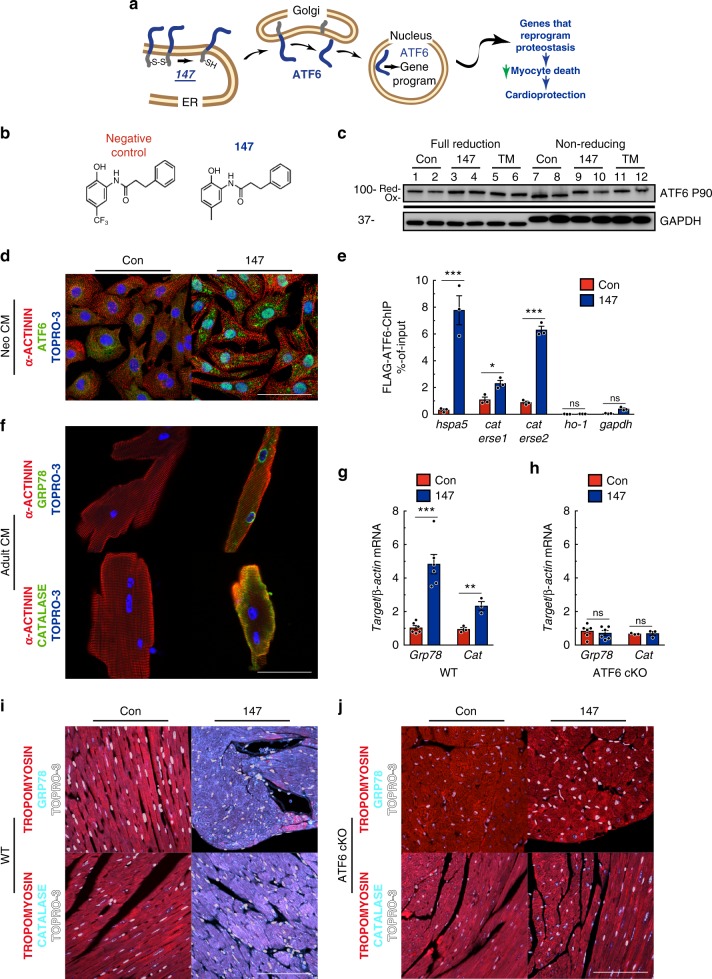
Fig. 3.
147 selectively activates ATF6 in the heart. a Diagram of the hypothetical mechanism of ATF6 activation by 147. b Chemical structure of synthetic control compound and compound 147. c Immunoblot of ATF6 and GAPDH in NRVM 24 hours after treatment with compound 147 or TM in fully reducing condition (lanes 1–6) or non-reducing conditions (lanes 7–12). Shift exhibited in Atf6 in TM-treated cells in full-reducing conditions is typical of de-glycosylated ATF6. Full uncut version of this gel can be found in Supplementary Figure 7. d Immunocytofluorescence (ICF) of ATF6 (green), α-actinin (red), and nuclei (TOPRO-3) in NRVM 24 hours after treatment with compound 147. Scale bar represents 50 µm. e Chromatin immunoprecipitation (ChIP-qPCR) of known ATF6 target promoter binding elements (ERSE) for Grp78 (hspa5), cat, and negative control targets Heme oxygenase 1 (ho-1) and gapdh NRVM infected with AdV encoding Flag-ATF6 (1–670) 24 hours after treatment with compound 147 (n = 3). f ICF of GRP78 and CAT (green), α-actinin (red), and nuclei (TOPRO-3) in AMVM 24 h after treatment with compound 147. g, h qPCR for Grp78 (n = 6) or Cat (n = 3) in LV of WT (g) or ATF6 cKO (h) hearts 24 hours post treatment with control or 147. i, j IHC staining of GRP78 or CAT (cyan), tropomyosin (red), and nuclei (TOPRO-3, white) in left ventricle (LV) of WT (i) or ATF6 cKO (j) hearts 24 hours post treatment with control or 147. Tissue sections are representative images from one mouse per condition. Scale bar represents 50 µm. Data are represented as mean ± s.e.m. Two-group comparisons were performed using Student’s two-tailed t test, and all multiple group comparisons were performed using a one-way ANOVA with a Newman–Keuls post hoc analysis. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001