Abstract
The CCCTC-binding factor (CTCF) is a central regulator of chromatin topology recently linked to neurodevelopmental disorders such as intellectual disability, autism, and schizophrenia. The aim of this study was to identify novel roles of CTCF in the developing mouse brain. We provide evidence that CTCF is required for the expression of the LIM homeodomain factor LHX6 involved in fate determination of cortical interneurons (CINs) that originate in the medial ganglionic eminence (MGE). Conditional Ctcf ablation in the MGE of mice of either sex leads to delayed tangential migration, abnormal distribution of CIN in the neocortex, a marked reduction of CINs expressing parvalbumin and somatostatin (Sst), and an increased number of MGE-derived cells expressing Lhx8 and other markers of basal forebrain projection neurons. Likewise, Ctcf-null MGE cells transplanted into the cortex of wild-type hosts generate fewer Sst-expressing CINs and exhibit lamination defects that are efficiently rescued upon reexpression of LHX6. Collectively, these data indicate that CTCF regulates the dichotomy between Lhx6 and Lhx8 to achieve correct specification and migration of MGE-derived CINs.
SIGNIFICANCE STATEMENT This work provides evidence that CCCTC-binding factor (CTCF) controls an early fate decision point in the generation of cortical interneurons mediated at least in part by Lhx6. Importantly, the abnormalities described could reflect early molecular and cellular events that contribute to human neurological disorders previously linked to CTCF, including schizophrenia, autism, and intellectual disability.
Keywords: cell transplantation, chromatin structure, CTCF, GABAergic interneurons, intellectual disability, schizophrenia
Introduction
The cerebral cortex consists of pyramidal excitatory neurons and inhibitory interneurons. The latter produce GABA to provide the rhythmic inhibitory activity that synchronizes pyramidal cell firing in mature neuronal circuits of the cortex (for review, see Tremblay et al., 2016). Not surprisingly, perturbation of the cortical GABAergic system underlies several neurodevelopmental disorders, including Down syndrome (Pérez-Cremades et al., 2010; Ruiz-Mejias et al., 2016), fragile X syndrome (Gibson et al., 2008; Paluszkiewicz et al., 2011), and nonsyndromic autism spectrum disorder (ASD) (Blatt and Fatemi, 2011; Hashemi et al., 2017), and pharmacological modulation of GABA receptors has been therapeutic in some contexts (Potier et al., 2014; Lozano et al., 2015). Recently, the transplantation of immature interneurons into the postnatal cortex has been explored as an alternative to pharmacological therapy in mouse models of epilepsy (Southwell et al., 2014; Hammad et al., 2015; Chohan and Moore, 2016). The laminar position and marker expression of cortical interneurons (CINs) depend more on intrinsic than extrinsic factors and transplanted cortical GABAergic interneurons have the potential to integrate into existing circuits. Understanding the developmental steps and regulatory events governing CIN development is thus essential to generating interneurons for successful therapeutic transplantations.
In mice, the majority of CINs are mainly produced in the ventral telencephalon in transient progenitor structures called the medial ganglionic eminence (MGE) and the caudal ganglionic eminence (CGE). MGE- and CGE-derived interneurons migrate tangentially into the developing cortical marginal and intermediate zones before radially invading the cortical plate and forming connections with the cortical pyramidal neurons (Marín, 2013). Transcriptional cascades in the MGE result in a peak of neurogenesis between embryonic days 12 (E12) to E14 that generates the somatostatin (Sst)- and parvalbumin (Pv)-expressing CIN subtypes (Xu et al., 2004; Miyoshi and Fishell, 2011). A subset of newly generated neurons and secondary progenitors located in the subventricular zone of the MGE express both Lhx6 and Lhx8, two LIM homeobox factors with partially overlapping function (Marin et al., 2000; Fragkouli et al., 2009). The cells destined to become CINs express only Lhx6, whereas the neurons that express Lhx8 (some in combination with Lhx6) are fated to become cholinergic or GABAergic neurons of the basal forebrain (Fragkouli et al., 2005, 2009). Lhx6-null interneurons resemble CGE-derived neurogliaform interneurons with regard to marker expression, laminar position, and electrophysiology (Vogt et al., 2014), indicating that LHX6 specifies the fate of MGE-derived CINs.
CCCTC-binding factor (CTCF) is a ubiquitously expressed protein that orchestrates loop formation in chromatin by binding to a consensus sequence and simultaneously dimerizing or partnering with other chromatin-associated proteins such as the cohesin complex (for review, see Merkenschlager and Nora, 2016). These loops can bring distant genomic elements such as promoters and enhancers into close proximity or may act as a barrier, such as when CTCF functions as an insulator protein. In parallel, emerging evidence suggests that CTCF is required for normal development and functioning of the CNS. De novo mutations in one copy of the human CTCF gene were recently identified in five patients with microcephaly, intellectual disability (ID), and short stature (Gregor et al., 2013; Bastaki et al., 2017). Two individuals with ID screened as part of a cohort selected for ASD were also identified as having mutations in one allele of CTCF (Iossifov et al., 2014). Polymorphisms in and around the CTCF gene have also been associated with schizophrenia in two different patient–control cohorts (Juraeva et al., 2014). Moreover, CTCF has been shown to regulate the expression of the protocadherin membrane adhesion proteins for which the combinatorial expression in neurons is thought to underlie the specificity of neuronal connectivity (Hirayama et al., 2012). The analysis of dorsal projection neurons in Ctcf-deficient mice revealed defects in dendritic arborization and reduced spine density (Hirayama et al., 2012). Viral-mediated knock-down of Ctcf in hippocampal cells also results in reduced spine density and leads to impaired learning (Sams et al., 2016).
In this study, we present our findings from Ctcf gene inactivation in the developing mouse ventral telencephalon. We show that CTCF expression is required for the normal development and fate specification of MGE-derived CINs. Ctcf deletion results in reduced expression of Lhx6 and an upregulation of Lhx8, resulting in aberrant fate specification and reduced number of PV+ and SST+ CIN subtypes. Reintroduction of Lhx6 in CTCF-null MGE progenitors is sufficient to reestablish the normal number of SST+ interneurons and lamination pattern in transplantation experiments. These findings suggest that loss of CTCF during telencephalon development can affect postnatal CIN number and therefore inhibitory functions, perhaps contributing to cognitive abnormalities.
Materials and Methods
Mice and genotyping.
Animal studies were approved by the University of Western Ontario Animal Care Committee according to guidelines established by the Canadian Council on Animal Care. Mice were housed in the London Regional Cancer Program vivarium (12 h light/dark cycle) and were provided with regular chow and water ad libitum. Ctcffl/fl mice were generated by Heath et al. (2008) on a C57BL/6 background. The NestinCre mice (C57BL/6 background) were described previously (Bérubé et al., 2005). Bbc3/Puma−/− (stock #011067), Nkx2.1Cre (stock #008661), and RosamTmG (stock #007576) mice were purchased from the The Jackson Laboratory. Mice carrying RosamTmG are of a mixed background and were bred to Ctcffl/fl mice to homozygosity.
Ctcffl/fl and NestinCre+/− mice were mated and the resultant Ctcffl/wt:NestinCre/− offspring were time mated to Ctcffl/fl mice to generate Ctcf conditional knock-out embryos (Ctcffl/fl; NestinCre+/−, hereafter referred to as CtcfNes-Cre), as described previously (Watson et al., 2014). CtcfNes-Cre embryos of either sex exhibit efficient Cre-mediated recombination at loxP sites flanking exons 3–12 of the Ctcf locus in neuroprogenitors beginning at approximately E11(Watson et al., 2014). Embryos of either sex without the NestinCre transgene and wild-type Ctcf expression were used as controls.
CtcfNkx-Cre embryos and mice were generated in the same fashion as described above for the CtcfNes-Cre mice. Nkx2.1Cre mice were maintained on a C57BL/6 background, with the exception of the transplantation experiments, which were performed using Nkx2.1Cre mice maintained on a CD1 background. For the mice used for transplantation experiments, care and procedures were performed according to the University of California at San Francisco Laboratory Animal Research Center guidelines.
For timed mating, time of conception was considered to be the midnight before vaginal plug discovery. To label proliferative cells, pregnant dams were injected intraperitoneally with BrdU labeling agent (GE Healthcare Life Sciences) at 1 ml/100 g body weight, as described previously, 1 h before being killed (Watson et al., 2014).
P20–P22 mouse brains were harvested by transcardial perfusion with 20 ml of PBS (DEPC-treated) followed by perfusion with 20 ml of 4% paraformaldehyde (PFA)/PBS (DEPC-treated) before brain dissection.
RNA-sequencing (RNA-Seq) analysis.
Telencephalons of two E14 CtcfNes-Cre embryos and littermate controls (Ctcf fl/ fl) were processed for RNA-Seq at Otogenetics. Raw reads were aligned to mouse genome mm10 using STAR (Dobin et al., 2013) version 2.5.3a with GENCODE M11 primary annotations. Count matrices were generated using the R GenomicAlignments package (Lawrence et al., 2013) and normalized using DeSeq2 (Love et al., 2014). Low-expressing genes (counts <2% of the average of all samples) were removed and genes with an average change of 1.5-fold or greater were used for downstream analysis by The Protein ANalysis THrough Evolutionary Relationships (PANTHER; http://pantherdb.org/).
qRT-PCR analysis.
For qRT-PCR, total RNA was extracted using an RNeasy Mini kit (Qiagen) and cDNA was synthesized using SuperScript II Reverse Transcriptase (Invitrogen), as described previously (Watson et al., 2014). Primer sequences used for qRT-PCR are listed in Table 1. All primers in this study were designed using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/primer3/). PCR amplification was performed using the iQ SYBR Green supermix (Bio-Rad) under the following conditions: 95°C for 3 min (]95°C for 10 s, 58°C for 20 s, 72°C for 30 s × 35) and 72°C for 5 min. A melting curve for each sample was generated from 55°C to 95°C in increments of 1°C and PCR products were resolved on agarose gels to verify band size. β-actin transcript was quantified and used for normalization of the data.
Table 1.
Primer sequences used for genotyping, qRT-PCR, and ISH probe synthesis
| Gene (purpose) | Forward sequence | Reverse sequence |
|---|---|---|
| Ctcf (genotype) | CTAGGAGTGTAGTTCAGTGAGGCC | GCTCTAAAGAAGGTTGTGAGTTC |
| Sry (genotype) | GCAGGTGGAAAAGCCTTACA | AAGCTTTGCTGGTTTTTGGA |
| Cre (genotype) | TGACCAGAGTCATCCTTAGCG | AATGCTTCTGTCCGTTTGCC |
| Puma (genotype) | AGGCTGTCCCTGCGGTCATCC | WT: GGACTGTCGCGGGCTAGACCCTCTA |
| MUT: ACCGCGGGCTCCGAGTAGC | ||
| Rosa mTmG (genotype) | CTCTGCTGCCTCCTGGCTTCT | WT: CGAGGCGGATCACAAGCAATA |
| MUT: TCAATGGGCGGGGGTCGTT | ||
| Ctcf (qRT-PCR) | CGATATGCTCTCATCCAGCA | TCCCACACTTGGAACAGACA |
| Lhx6 (qRT-PCR) | CTTGGGCTGACTGTCCTGTT | GGTGCGGCAGACAAATCTAT |
| B actin (qRT-PCR) | CTGTCGAGTCGCGTCCACCC | ACATGCCGGAGCCGTTGTCG |
| Lhx8 (qRT-PCR) | CCACCCATGTTGGAAGAAAT | CATTGGATGGGGTAACAAGG |
| Cxcr4 (qRT-PCR) | GAAGTAGATGGTGGGCAGGA | ACGGCTGTAGAGCGAGTGTT |
| Sst (qRT-PCR) | GGGCATCATTCTCTGTCTGG | GAGGCAAGGAAGATGCTGTC |
| Lhx8 (probe) | GAAGAGCGATCAGATGTTTGTG | TAATACGACTCACTATAGGGCACCTGTATGACACGTCTGCTT |
| Pvalb (probe) | GGATGTCGATGACAGACGTG | TAATACGACTCACTATAGGGACTCAACCCCTTCCCTTCC |
| Sst (probe) | TGAAGGAGACGCTACCGAAG | TAATACGACTCACTATAGGGAGGGTCAAGTTGAGCATCG |
| Gbx2 (probe) | GATGAAGAGAAGCTAGAGCCCC | TAATACGACTCACTATAGGGAGGTTCAGATCCTGTGACTTCC |
| Gad1 (probe) | GGGACCCTTGAACCGTAGAG | TAATACGACTCACTATAGGGCCCGTACTTCAGGGTGTCTC |
| Cr (probe) | GATGCTGACGGAAATGGG | TAATACGACTCACTATAGGGCCCTACCAGCCACCCTCT |
| VIP (probe) | CCTGGCATTCCTGATACTCTTC | TAATACGACTCACTATAGGGATTCTCTGATTTCAGCTCTGCC |
Immunofluorescence (IF) and in situ hybridization (ISH) staining.
Dissected samples were incubated in 4% PFA/PBS (DEPC-treated) overnight and then dehydrated in 30% sucrose until samples sank to the bottom before being frozen in Cryomatrix (optimal cutting temperature medium) and sectioned at 8 μm thickness. IF and BrdU labeling was performed as described previously (Watson et al., 2014). For IF, slides with sections were rehydrated in PBS and then incubated overnight (8–16 h) with the primary antibody at 4°C in 0.3% Triton X-100 in PBS at the following dilutions: rabbit anti-CTCF (1:400; Cell Signaling Technology), mouse anti-BrdU (1:50; BD Biosciences), rabbit anti-Nkx2.1 (1:100; Santa Cruz Biotechnology), and goat anti-SP8 (1:100; Santa Cruz Biotechnology). For BrdU, tissue sections were treated with 2 n HCl and then 0.1 m Na2B4O7, pH 8.5, before primary antibody incubation. For anti-SP8 staining, slides were heated in 10 mm sodium citrate and then blocked with 10% NGS/0.3% Triton X-100 in PBS for 1 h before incubation with the primary antibody. The following morning, sections were washed with 0.3% Triton X-100 in PBS and incubated with the corresponding secondary antibody (1:800 Alexa Fluor 594 or 488; Invitrogen) for 1 h. Sections were then washed again and stained with DAPI (Sigma-Aldrich).
ISH riboprobes were synthesized as follows, with the exception of the Reelin ISH probe, which was a kind gift from the Rubenstein laboratory; Template DNA was PCR amplified using primers; the reverse primer was labeled with the T7 recognition sequence at the 5′ end (Table 2). Digoxigenin-labeled riboprobes were then synthesized from the gel-purified template DNA using T7 (antisense probe) polymerase (Roche) according to the manufacturer's instructions. For ISH, sections were incubated overnight (12–18 h) with antisense digoxigenin-labeled riboprobes at 65°C in a humidified chamber. Riboprobes were diluted in formamide-based hybridization buffer at 1:1000 from stock. The following morning, sections were washed in formamide-based ISH wash buffer and then maleic acid buffer containing Tween 20 (MABT) before incubation with alkaline phosphatase conjugated anti-figoxigenin Fab fragments (Roche) overnight. Excess antibody was removed with a series of MABT and prestaining buffer washes before riboprobe detection using the NBT/BCIP system (Roche) in a polyvinyl alcohol-based solution. After staining, the reaction was ended with vigorous PBS washing. Sections were cleared by dehydration in serial ethanol dilutions (70%, 90%, and 100%) and xylene and then mounted.
Table 2.
Number of GFP+ cells observed 35 d after MGE cell transplantation for each genotype
| Transplanted MGE cells | n | No. of transplanted cells | No. of brain sections assayed | No. of cells/section | Mean no. of cells/section | SEM |
|---|---|---|---|---|---|---|
| CTCF cHet | 1st | 219 | 42 | 5.21 | 8.68 | 1.88 |
| 2nd | 490 | 42 | 11.67 | |||
| 3rd | 357 | 39 | 9.15 | |||
| CTCF cKO | 1st | 76 | 36 | 2.11 | 4.65 | 1.29 |
| 2nd | 248 | 45 | 5.51 | |||
| 3rd | 266 | 42 | 6.33 | |||
| CTCF cKO/Lhx6 rescue | 1st | 161 | 24 | 6.71 | 4.79 | 0.96 |
| 2nd | 88 | 24 | 3.67 | |||
| 3rd | 96 | 24 | 4.00 |
Microscopy and imaging.
Fluorescence and light microscopy was performed using an inverted (DMI 6000b) Leica microscope. Light microscopy was also performed with the Aperio CS2 digital pathology scanner (Scanscope; Leica). For transplantation experiments, fluorescent images were taken using a Coolsnap camera (Photometrics) mounted on a Nikon Eclipse 80i microscope using NIS Elements acquisition software (Nikon). Images were processed with Volocity (PerkinElmer), Imagescope (Leica), ImageJ, and Adobe Photoshop.
Coimmunoprecipitation and Western blot analysis.
Total protein was extracted from embryonic telencephalon or MGE using standard protocol with RIPA buffer and quantified with the Bradford assay. For immunoprecipitation, E13.5 MGEs were pooled from wild-type embryos and the protein was incubated overnight with anti-CTCF, anti-NKX2.1, or IgG antibody (mock IP). Antibody–protein complexes were incubated with protein-G Dynabeads and isolated by centrifugation. After washing, interactions were disrupted and proteins were detected by Western blotting as described below.
Protein lysates were resolved on an 8% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. Anti-CTCF (1:1000; Cell Signaling Technology), anti-NKX2.1 (1:2000; Santa Cruz Biotechnology), and anti-β-Actin (1:7000; Sigma-Aldrich) were used to probe the membrane, followed by secondary detection with the corresponding horseradish peroxidase-conjugated antibodies (1:4000; GE Healthcare Life Sciences). Protein was quantified using ImageJ software (version 1.47).
ChIP sequencing.
ChIP in MGE was performed as described previously for mouse forebrain (Watson et al., 2014). E13.5 MGE was dissected and pipetted up and down in DMEM to create a single-cell suspension. Cells were fixed in 1% formaldehyde and then lysed in SDS buffer. Chromatin was sonicated to an average size of 500 bp and then 1/25 of total chromatin was set aside (input). The rest of the chromatin was incubated overnight with rabbit anti-CTCF antibody (Cell Signaling Technology) or rabbit IgG as a control. CTCF-bound chromatin fragments were then purified using magnetic Dynabeads (Invitrogen) and crosslinking was reversed. ChIP-sequencing (ChIP-Seq) reads were aligned to mouse genome mm9 or mm10 using Bowtie2 (Langmead and Salzberg, 2012) version 2.3.2 using default settings. Peaks were called using MACS2 (Zhang et al., 2008) version 2.1.1.20160309 with p = 0.001. Wig files were generated using deepTools (Ramírez et al., 2014) and overlapping peaks were identified using Bedtools (Quinlan and Hall, 2010).
T4 ELISA.
Blood was collected from P20 mice by transcardiac puncture using an EDTA-coated syringe, pH 7.0. Blood was centrifuged for 10 min (14,000 RPM) at 4°C and then the plasma collected and stored at −20°C. Plasma was loaded onto the T4 ELISA kit (T4044T-100; Calbiotech) according to manufacturer's protocols.
Cell counts.
To determine the density of marker-positive cells in P20–P22 cortex, results from a representative rectangle in somatosensory cortex from 3–6 serial cryosections (8 μm thickness) per animal were averaged. Because the number of transplanted cells is low, tissue section thickness of 25 μm was used to ensure adequate power to detect differences between test and control samples for transplantation experiments.
The cortical rectangle was ∼400 μm wide and depth was determined by inclusion of all cortical layers. Areas within the indicated brain regions were calculated (in square millimeters) from microscope images in Volocity or Imagescope software and counts were performed in a manner blinded to genotype. For Gad1 and Reelin marker-positive cell counts, the cortex was divided into 10 equal-sized horizontal bins to simultaneously determine laminar changes. The first cortical bin location and size corresponds approximately to the marginal zone.
For cell counts in the MGE, images were acquired at a magnification of 20× to allow for sufficient area and visual quality. To determine cell densities for BrdU+, ki67+, and PH3+ cells at E14, standard parameters were set for a free-hand-generated shape. The shape edges followed the contour of the MGE at ∼100 μm wide and the depth of the MGE ventricular zone at ∼100 μm deep.
Cloning.
The DlxI12b-GFP-T2a-Lhx6 vector (Vogt et al., 2014) was modified to remove loxP sites that flanked the expression cassettes. PCR was used to generate a DlxI12b-GFP fragment that harbored a 5′ XbaI restriction enzyme site, but excluded the loxP site from the original vector and a 3′ primer against GFP that included a BsrGI site. This product was then ligated into the same backbone in the 5′ XbaI and 3′ BsrGI sites. The resulting vector lacked a flanking loxP site and could be expressed in cells expressing Cre recombinase. Vectors were verified by restriction digest and sequencing.
Lentivirus generation.
HEK293T cells grown in DMEM H21 with 10% FBS were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) with the lentiviral expression vector and three helper plasmids (pVSV-g, pRSVr, and pMDLg-pRRE) to generate lentivirus particles as described previously (Vogt et al., 2015). Media containing lentiviruses was collected after 4 d and filtered through a 0.45 low-protein-binding membrane to remove cells and large debris. The filtered medium was pooled and ultracentrifuged at 100,000 × g for 2.5 h at 4°C. The supernatant was removed and the pellet was resuspended overnight at 4°C in sterile PBS and then stored at −80°C until use.
MGE transplantation.
MGE transplantation and rescue assays have been described previously (Vogt et al., 2014). Briefly, E13.5 MGE tissue from either Ctcf flox/wt:NestinCre+ (controls) or Ctcf flox/flox:NestinCre+ (knock-out) embryos were harvested and dissociated into a single-cell suspension before injection into a wild-type P1 host. For rescue experiments, MGE cells from Ctcf flox/flox:NestinCre+ embryos were collected in the same manner and then incubated with lentiviruses that expressed Lhx6 and Gfp for 30 min at 37°C in media at physiological pH before transplantation. Transplanted cells were allowed to develop in vivo for 35 d before analysis. Immunofluorescence staining of sections at day 35 were performed with the following antibodies: rabbit anti-GFP (Clontech, 1:2000), chicken anti-GFP (Aves Laboratories, 1:2000), rabbit anti-PV (Swant, 1:500), mouse anti-PV (Millipore, 1:500), rat anti-SST (Millipore, 1:200), or guinea pig anti-LHX8 (a generous gift from Aleksandar Rajkovic, University of Pittsburgh).
Experimental design and statistical analysis.
Sample sizes of test animals were chosen based on the minimum number of animals required, in our experience, to detect differences between control and experimental samples. To minimize within-pair variation, we used littermate-matched, sex-matched animals. Because we have detected no difference in phenotype between male and female animals in past or present study of this animal model, results from both sexes were combined.
Given our previous work in the NesCre Ctcf model, transcriptional changes in the absence of CTCF were investigated at the E13.5–E14.5 developmental age, a time before onset of cell death. Given the early, highly penetrant lethality of NesCre-mediated deletion of Ctcf, we selected a new Cre driver line for specific deletion of Ctcf from the MGE. Nkx2.1Cre mice were selected because this model is well characterized, with early Cre expression and limited off-target effects, which include Cre-mediated deletion in lung and thyroid tissues. The E15.5–E16.5 time point was selected to analyze migration of MGE-derived interneurons based on previous studies showing that a large proportion of MGE-derived interneurons have completed tangential migration by this time. After birth, animals were weighed daily and survival was tracked. The P20–P22 time point was selected based on animal survival and for comparison with the study by Vogt et al. (2014) and the histological data presented from this time point were generated from six conditional knock-outs and their control littermates (two female pairs and four male pairs).
Tissue sections were taken in series (10 sections apart), as opposed to sequentially. For cell density counts, one slide with five series sections was stained per marker per animal. Given some sections were not of appropriate quality, this allowed for three to five sections to be photographed and used for quantification. A representative image was selected among these. Counting was done by an individual blinded to genotype. Markers for this study were selected based on careful review of the literature. Gapdh and β-actin were used for normalization based on their established use as controls and previous use in these models. For qRT-PCR of Lhx6, Lhx8, Sst, and Gbx2, three pairs of control and knock-out animals were used. For cell counts of BrdU, Ki67, and PH3, three pairs were used. For cell counts of interneuron markers, sample sizes were as follows: Gad1, n = 3; Reelin, n = 3; Vip, n = 3; Cr, n = 4; Pv, n = 4; Sst, n = 3; and SP8, n = 3. All other sample sizes are listed in the figure legends. We analyzed qRT-PCR, Western blot, ELISA, and cell counts with two-tailed Student's t tests. Because Gad1 and Reelin are interneuron markers that are present across multiple laminar layers, the cortex was divided into bins for these markers to investigate lamination. The bins were equal in size for ease and consistency between samples and the number 10 was selected as then the width of “bin 1” would approximate the marginal zone layer, which was of particular interest based on the Vogt et al. (2014) study. Repeated-measures ANOVA was used for evaluation of the binned data across the cortex.
Given the association of transcriptional changes in Lhx6 with the Ctcf mutant phenotype, we designed a rescue experiment to demonstrate a causal link. The reexpression of Lhx6 using lentiviral construct was chosen due to the previous success of this method and the fact that it eliminates confounding effects of tissue-autonomous Ctcf deletion and we could thus investigate Ctcf-null MGE cells in a wild-type tissue environment. In addition, this method eliminated the potential confounder of impaired tangential migration because cells are transplanted directly into the cortical milieu.
Results
Altered expression GABAergic neuron differentiation genes in the CTCF-null telencephalon
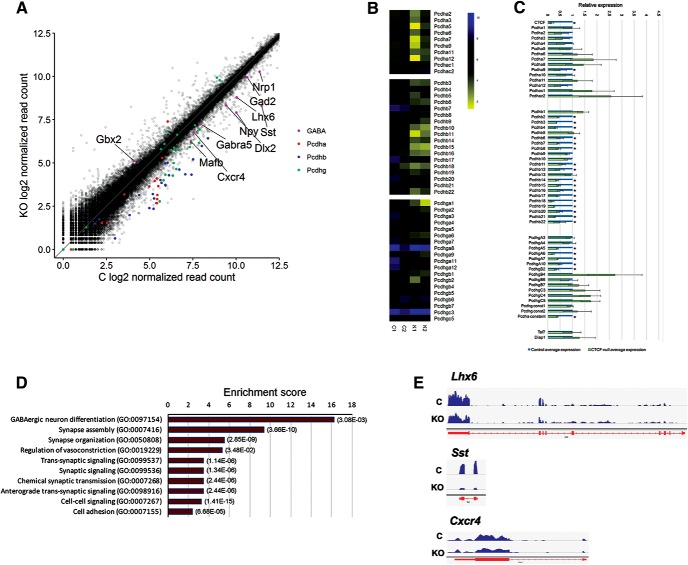
We previously reported telencephalon defects upon conditional inactivation of Ctcf in the embryonic brain using the NestinCre driver line of mice (Watson et al., 2014), which deletes Ctcf in early neural progenitors (radial glia). Analysis of the dorsal forebrain of E16.5 CtcfNes-Cre revealed extensive Tp53- and Puma-dependent cell death and premature differentiation of neural precursor cells, leading to depletion of the progenitor pool (Watson et al., 2014). To determine the effects of Ctcf inactivation on gene expression, we performed RNA sequencing of CtcfNesCre and littermate control telencephalon at E14 before apoptosis induction. Using the criteria of a minimum expression of 2% of the average reads and a minimum average fold change of 1.5, we generated a list of genes (N = 2024) with altered expression in the CtcfNes-Cre E14 telencephalon. Approximately half of these genes (n = 1050) exhibited decreased expression and included a large number of the clustered Ca2+-binding protocadherin (Pcdh) genes, with a more pronounced effect on Pcdh-β genes (Fig. 1A,B). We confirmed the effect of Ctcf inactivation on Pcdh genes by qRT-PCR (Fig. 1C). These results expand on previous reports by demonstrating that CTCF regulates Pcdh cluster genes in the embryonic brain before neuronal network establishment. However, we found that many genes displaying expression differences in the CtcfNes-Cre E14 telencephalon were involved in the early development and differentiation of GABAergic interneurons (Fig. 1A). Indeed, the most significantly enriched Gene Ontology (GO) Biological Processes term with >2.0 fold enrichment (PANTHER version 11.1, Pantherdb.org) was GABAergic neuron differentiation (Fig. 1D), suggesting a potential novel role of CTCF in the development of inhibitory interneurons. The list of genes included Lhx6, an important upstream regulator of GABAergic neuron fate specification (Liodis et al., 2007), and several Lhx6-dependent genes or downstream effectors, including Sst and Cxcr4 (Fig. 1E). Additional GO enrichment terms were related to synaptic transmission, which is expected from the large number of altered Pcdh genes and recent studies implicating CTCF in learning and memory (Sams et al., 2016).
Figure 1.
Transcriptional profiling of CTCF-null and control E14 telencephalon. A, Log2 of normalized read counts of control and CtcfNes-Cre E14 telencephalon RNA-Seq. Genes implicated in GABAergic differentiation and Pcdh cluster genes are highlighted in color. B, Heat maps of Pcdh gene expression across the α, β, and γ clusters from RNA-Seq analysis. C, qRT-PCR of Pcdh genes from clusters α, β, and γ in CtcfNes-Cre normalized to littermate controls and to Gapdh. n = 3. Error bars indicate SEM. Asterisks indicate p < 0.05 using Student's t test, not corrected for multiple testing. Taf7 and Diap1 are genes located between the Pcdh-β and Pcdh-γ clusters. D, Top 10 enriched GO terms for biological processes of genes downregulated in the CtcfNes-Cre E14 telencephalon. The list was generated using the PANTHER overrepresentation test. p-values with a Bonferroni correction are shown on the right for each category. E, University of California Santa Cruz genome browser view of RNA-Seq data of selected genes involved in GABAergic interneuron differentiation.
Ctcf regulates the expression of Lhx6 and Lhx8 in the MGE
Lhx6 is required in the developing MGE for the production of PV+ and SST+ interneuron subtypes that ultimately populate multiple areas of the forebrain, including the striatum and cerebral cortex. qRT-PCR confirmed an ∼2.5-fold decrease in Lhx6 transcript levels (p = 0.028). Similarly, the expression of Sst, a known downstream target of LHX6, exhibits an ∼10-fold decrease (p = 0.0001) (Fig. 2A). ISH of E13.5 embryonic sections confirmed decreased Lhx6 mRNA and a reduced number of Sst+ cells in the CtcfNes-Cre MGE and forebrain, respectively (Fig. 2B). We observed that, in a subset of embryos, the reduction in Lhx6 expression was restricted to the caudal MGE (data not shown). ISH of Sst transcripts identified fewer Sst+ cells migrating tangentially from the MGE toward the cortex in the CtcfNes-Cre compared with control embryos (Fig. 2C). Given that CTCF protein expression is not detectable in the MGE of the CtcfNes-Cre embryos by E12.5 (Watson et al., 2014; Fig. 2F), we examined Lhx6 expression at this time point by ISH and observed only a subtle decrease in the CtcfNes-Cre MGE compared with control (Fig. 2G), suggesting that Lhx6 expression is affected in the CtcfNes-Cre MGE mostly after E12.5 and this reduction is most pronounced in cells of the posterior MGE.
Figure 2.
Fewer Lhx6+ and Sst+ cells are present in the CTCF mutant embryonic brain. A, qRT-PCR performed in E14 telencephalon (or MGE) isolated from control (gray) or CtcfNes-Cre (blue) embryos (n = 3 pairs). Data were normalized to β-actin expression. Asterisks indicate p < 0.05, Student's t test. Error bars indicate SEM. ISH of the indicated transcripts on coronal brain sections of E13.5 control and CtcfNes-Cre embryos (B, C) or PUMA−/− and CtcfNes-Cre/PUMA−/− embryos (D, E). F, CTCF immunofluorescence staining and DAPI counterstain in E12.5 CtcfNes-Cre brain coronal sections (Watson et al., 2014). Adjacent insets show that CTCF is expressed in the MGE in control, but absent in the CtcfNes-Cre MGE, at E12.5. G, Lhx6 ISH of E12.5 rostral and caudal coronal sections. H, BrdU immunofluorescence staining of embryonic MGE section 1 h after BrdU injection of pregnant dam. I–K, Quantification of BrdU+ (I), Ki67+ (J), and phosphohistone H3 (PH3+) (K) nuclei per 100 mm2 in E12.5 MGE VZ of control and CtcfNes-Cre embryos (n = 3). Error bars indicate SEM. Scale bars, 100 μm. NS, Not significant.
To eliminate the possibility that the observed reduction in Lhx6 transcripts could be a consequence of Ctcf-null neuroprogenitor cell death, we assessed Lhx6 and Sst expression in CtcfNes-Cre embryos in a PUMA-null background. We found that preventing apoptosis of cells in the CtcfNes-Cre telencephalon via removal of PUMA failed to rescue the level of Lhx6 or Sst (Fig. 2D,E). We next addressed whether the reduction of Lhx6 and Sst transcript levels in Ctcf mutant embryos is caused by altered proliferation. Acute BrdU incorporation assays showed no significant difference in the proliferative capacity of the Ctcf-null neuroprogenitors in the ventricular zone of the MGE at E12.5 (Fig. 2H,I). Quantification of Ki67+ and phosphohistone H3+ (PH3+) cells in the MGE confirmed that proliferation is not affected (Fig. 2J,K). Together, these results indicate that the reduction in Lhx6 and Sst transcripts is not caused by altered survival or proliferative capacity of Ctcf-deficient MGE cells.
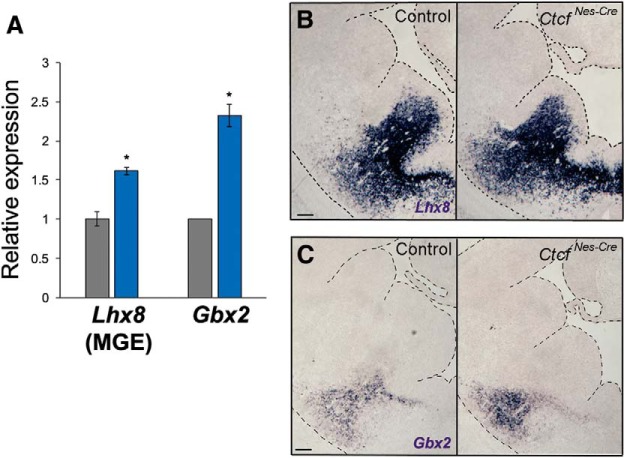
Beginning at E9.75, MGE cells are specified in the ventricular zone by Nkx2.1, a transcription factor with MGE-restricted expression that regulates the expression of Lhx6 and Lhx8. Through unknown mechanisms, a subset of the cells that coexpress these genes become either Lhx6+ or Lhx8+ (Fragkouli et al., 2009). Based on findings in Lhx8-deficient mice, LHX8 promotes a genetic program specifying telencephalic cholinergic interneurons that remain in the basal ganglia as striatal interneurons or projection neurons of regions such as the globus pallidus, ventral pallidum, and septum (Zhao et al., 2003). We therefore also examined the effect of CTCF loss on Lhx8 expression by ISH and qRT-PCR of RNA isolated from the MGE. In contrast to Lhx6, the expression of Lhx8 was significantly increased compared with control (p = 0.0058), suggesting that a greater number of MGE cells retain Lhx8 at the expense of Lhx6 expression (Fig. 2A,B). Increased expression of Gbx2, a downstream target of Lhx8 (Zhao et al., 2003), supports a scenario in which Ctcf-null MGE cells might implement different fate pathways due to reduced Lhx6 expression and increased Lhx8 expression (Fig. 3A–C).
Figure 3.
Increased expression of Lhx8 and Gbx2 in the Ctcf-null MGE. A, qRT-PCR of telencephalon (or MGE) isolated from control (gray) or CtcfNes-Cre (blue) embryos (n = 3 pairs). Data were normalized to β-actin expression. Asterisks indicate p < 0.05, Student's t test. Error bars indicate SEM. B, C, ISH of Lhx8 (B) and Gbx2 (C) performed on coronal brain sections of control and CtcfNes-Cre E13.5 embryos. Scale bars, 100 μm. Tissue edges are outlined for ease of visualization.
CTCF acts independently of NKX2.1 to regulate Lhx6 and Lhx8 expression
Given that NKX2.1 induces the expression of Lhx6 and Lhx8, we investigated whether CTCF affects these factors by controlling NKX2.1 expression in the embryonic MGE, but found no evidence that this is the case. Western blot analysis demonstrated that NKX2.1 is not altered in E13.5 CtcfNes-Cre compared with littermate control forebrain (Fig. 4A). We also stained coronal slices of E13.5 CtcfNes-Cre and littermate control embryos with an anti-NKX2.1 antibody, revealing no gross change in NKX2.1 levels or spatial expression (Fig. 4B). We next tested whether CTCF associates with NKX2.1 to regulate Lhx6 and Lhx8 by coimmunoprecipitation of CTCF and NKX2.1 in pooled MGE tissue dissected from wild-type E13.5 embryos and observed no interaction between these two proteins (Fig. 4C). In addition, we compared the chromatin occupancy of these two factors by ChIP-Seq analysis. We performed ChIP-Seq for CTCF using pooled wild-type E13.5 MGE tissue and the data obtained were compared with previously published NKX2.1 ChIP-seq data (Sandberg et al., 2016). We found that only a small percentage of CTCF peaks (6.5%) overlapped with NKX2.1-binding sites (p < 0.01, Fig. 4D). Regulatory elements previously identified in the promoter and in the first intron of Lhx6 (Du et al., 2008; Chen et al., 2013) are not bound by CTCF (Fig. 4E). We also examined CTCF binding to several enhancers that have activity in the MGE (Chen et al., 2013): (1) enhancer 422 [chr2:71,373,435-71,374,614 (mm9)], (2) DlxI12b [chr2:71,374,047-71,374,552 (mm9)], (3) the murine sequence corresponding to enhancer 692 near the human SOX6 gene [chr7:122,275,029–122,275,957 (mm9)], (4) the murine sequence corresponding to the human enhancer 1056 [chr18:81,349,730-81,351,184 (mm9)], and (5) the murine sequence corresponding to the human enhancer 1538 [chr2:71,373,514-71,374,608 (mm9)]. No peaks corresponding to CTCF binding were found at any of these sites (data not shown). Conversely, we observed CTCF-binding sites flanking a large genomic region surrounding Lhx6, suggesting that CTCF may promote long-range loop formation that is perhaps required for proper regulation of Lhx6.
Figure 4.
CTCF functions independently of Nkx2.1 to regulate Lhx6 and Lhx8. A, Western blot of Nkx2.1 in E13.5 telencephalon of controls and CtcfNes-Cre (KO) embryos. β-actin was used for normalization. Quantification is shown graphically on the right. Error bars indicate SEM. p = 0.968, Student's t test. B, Nkx2.1 immunofluorescence staining (red) in E13.5 coronal sections. Nuclei were counterstained with DAPI (blue). Scale bar, 100 μm. C, Co-IP of CTCF and Nkx2.1 of pooled wild-type MGE tissue. Asterisks indicate nonspecific IgG band. D, Venn diagram showing the overlap of CTCF and Nkx2.1 ChIP-Seq peaks in the MGE (p < 0.01). E, Diagram depicting UCSC genome browser views of the Lhx6 and Lhx8 loci (boxed), ChIP-Seq peaks for Nkx2.1, CTCF and input as well as the RNA-Seq tracks of control and CtcfNes-Cre E14 telencephalon. Key CTCF-binding sites are indicated by arrows.
We also examined CTCF binding at the Lhx8 locus and identified a peak in intron 6 and another ∼30 kb upstream of the gene (Fig. 4E). This pattern of CTCF binding has been observed at other loci that exhibit negative transcriptional regulation by CTCF, including the Bbc3/PUMA gene (Gomes and Espinosa, 2010). Altogether, these data suggest that CTCF does not affect NKX2.1 levels, but rather acts in parallel (potentially by altering chromatin topology) to control Lhx6 and Lhx8 expression.
MGE-specific deletion of Ctcf diminishes the number of Pv+ and Sst+ CINs
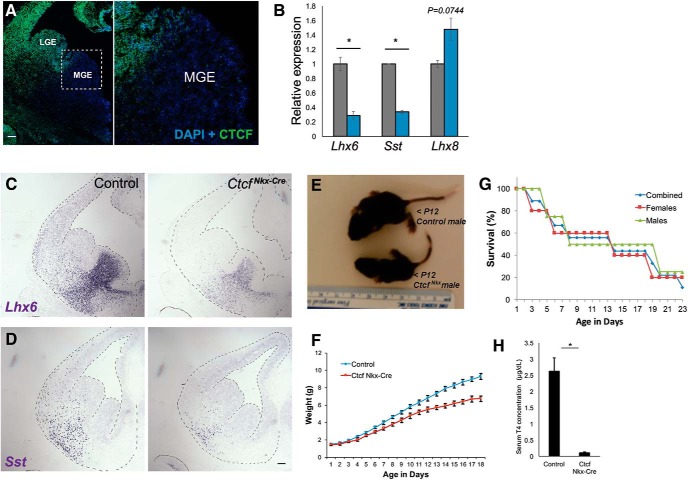
Although useful for the investigation of early forebrain development, the CtcfNes-Cre mice have some limitations. Extensive cell death ensues beyond E16 in the telencephalon and the mice die in the perinatal period, preventing any postnatal analysis of CINs. To circumvent these difficulties, we conditionally deleted Ctcf in the MGE starting at E10 with the Nkx2.1Cre driver line of mice. Immunofluorescence staining of E13.5 sections confirmed an absence of CTCF expression in most of the MGE, except for the dorsomedial MGE in some embryos, as reported previously (Vogt et al., 2014; Fig. 5A). Importantly, Ctcf deletion in the E13.5 MGE recapitulates differences in Lhx6, Sst, and Lhx8 transcript levels detected in CtcfNes-Cre embryos by qRT-PCR and ISH (Fig. 5B–D). CtcfNkx-Cre mice are smaller than controls and exhibit failure to thrive postnatally (Fig. 5E–G). Although this could be caused by the loss of CTCF in the MGE, Nkx2.1-driven Cre expression can sometimes occur in the lung and the thyroid (Minoo et al., 1999), so reduced survival of CtcfNkx-Cre might be caused by Cre-mediated deletion of Ctcf in these tissues. In support of this, we found that circulating thyroxine (T4) levels were reduced in CtcfNkx-Cre mice aged P20–P22 (Fig. 5H).
Figure 5.
MGE-specific Ctcf deletion replicates gene expression changes and extends postnatal survival. A, CTCF immunofluorescence (green) of E13.5 CtcfNkx-Cre coronal brain sections. Nuclei are counterstained with DAPI. Higher-magnification view of the MGE is shown on the right. Scale bar, 100 mm. B, qRT-PCR of Lhx6, Sst, and Lhx8 in control (gray) and CtcfNkx-Cre (blue) E14 telencephalon normalized to β-actin (n = 3). C, D, ISH of Lhx6 and Sst in E13.5 coronal sections. Scale bar, 100 mm. Tissue is outlined for ease of visualization. E, Representative image of a 12-d-old CtcfNkx-Cre male mouse and littermate control. F, Weight of CtcfNkx-Cre (n = 4) and littermate controls (n = 12) over time. Error bars indicate SEM. G, Survival curve of CtcfNkx-Cre mice (males = 4; females = 5). Five animals were killed due to severe distress or imminent death. H, Quantification of serum T4 from P20–P22 CtcfNkx-Cre mice and control siblings by ELISA. Error bars indicate SEM. n = 6 pairs. Asterisk indicates p < 0.05 by Student's t test.
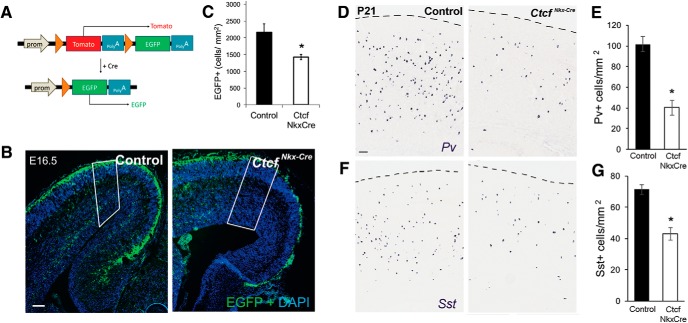
Tangential migration of MGE interneurons into the developing dorsal pallium is mediated by several molecular signals downstream of LHX6, including the interaction of chemoreceptors present on CIN cell surface (CXCR4 and CXCR7), with stromal cell-derived factor 1 (SDF1), which is present in the marginal and intermediate zones of the developing neocortex (Abe et al., 2015). To test for defects in MGE-derived interneuron migration in the CtcfNkx-Cre embryos, we introduced the Cre-responsive Rosa-mTmG reporter gene that results in GFP expression upon Cre-mediated recombination (Fig. 6A). The number of GFP+ MGE-derived cells were identified and counted in the E16.5 cortex of CtcfNkx-Cre and littermate control embryos. There was a significant decrease in the number of GFP+ cells in the CtcfNkx-Cre neocortex, consistent with a defect in tangential migration to the cortex (Fig. 6B,C). In support of a migration defect, the expression of Cxcr4 was significantly decreased in the CtcfNkx-Cre and CtcfNes-Cre E14 embryos compared with controls (data not shown).
Figure 6.
Reduced number of Sst- and Pv-expressing CINs in CtcfNkx-Cre mice. A, Diagram depicting the Rosa-mTmG allele and the Cre-dependent recombination that results in a switch from red to green fluorescence. Orange arrows represent loxP sites. B, Fluorescence microscopy of E16.5 cortical sections of the indicated genotypes after DAPI counterstaining (blue). Green fluorescence is evident in Cre+ cells originating from the MGE that are migrating tangentially in the cortex. Note that the red fluorescence channel is not shown. Scale bar,100 μm. Nkx-Cre control animals were used for this experiment. C, Quantification of GFP+ cells (outlined in white in B), n = 5 controls, n = 6 CtcfNkx-Cre. D, F, Pv and Sst ISH in the cortex of P21 CtcfNkx-Cre mice and littermate controls. E, G, Quantification of Pv+ cells (n = 4 pairs) or Sst+ cells (n = 3 pairs). Error bars indicate SEM and asterisks indicate p < 0.05, Student's t test.
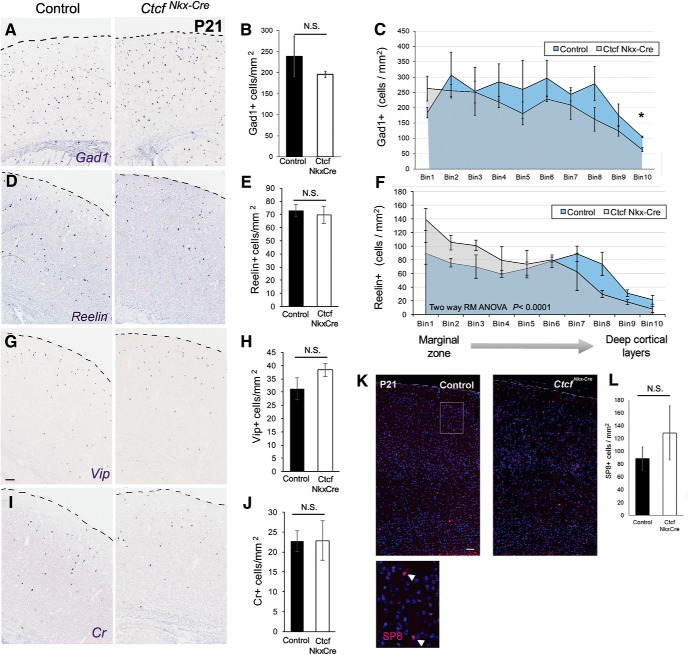
The extended viability of these mice compared with the CtcfNes-Cre allowed an investigation of CINs at P21, a time when CIN number and laminar position have attained adult patterns (Del Rio et al., 1992). ISH of brain sections from control and mutant mice revealed that the number of Pv+ and Sst+ interneurons in the CtcfNkx-Cre cortex is significantly reduced compared with controls (Fig. 6D–G). We also examined the outcome of CTCF loss on laminar positioning of interneurons by dividing a sampled cortical area into 10 equal bins, with bin 1 corresponding to the most superficial of these (i.e., cortical layer I). This analysis showed that Sst+ and Pv+ cells in CtcfNkx-Cre cortices were reduced equally among lamina (data not shown). Because MGE-derived interneurons occupy laminar layers based on developmental time, these findings suggest that Ctcf deletion affects MGE interneuron production equally over time.
Deletion of Ctcf results in altered fate and laminar distribution of MGE-derived cells
To further evaluate the postnatal consequences of the transcriptional alterations of Lhx6 in the embryonic CTCF-null MGE, we investigated the relative proportion of interneuron subtypes, as well as their respective cortical position, at P21. Despite the observed reduction of Pv+ and Sst+ cells in the cortex, the total number of CINs (identified by Gad1 staining) was not significantly different (Fig. 7A,B). Moreover, we observed no difference in the number of Reelin+ interneurons, a gene expressed in a portion of interneurons originating from the MGE and the CGE (Fig. 7D,E). However, Reelin+ interneurons in the CtcfNkx-Cre cortex tend to occupy upper cortical layers at the expense of deeper ones compared with controls (Fig. 7D,F).
Figure 7.
CTCF is required for CIN distribution across cortical layers. A, D, G, I, ISH of P21 cortices with the indicated probes. B, E, H, J, Quantification of cortical cell densities. Error bars indicate mean ± SEM. C, F, The cortex was divided into 10 horizontal bins, where Bin 1 represents the most superficial layer. Two-way repeated-measures ANOVA was significant for both Gad1 and Reelin. Student's t test was performed for each bin and significantly different bins (p < 0.05) are marked by asterisks (not corrected for multiple testing). K, Immunofluorescence staining for SP8 (red) in P21 cortex counterstained with DAPI. Merged blue and red channels are shown. Scale bar, 100 μm. Bottom inset shows higher magnification of SP8+ cells. L, Quantification of SP8+ cells. Error bars indicate mean ± SEM, n = 3 pairs. p = 0.432, Student's t test.
Because CGE-derived interneurons tend to occupy superficial layers, we hypothesized that there was aberrant specification of the MGE-derived cells into CGE-like cells. This possibility is supported by previous work showing that Lhx6 deletion causes a fate switch to more CGE-like subtypes as determined by laminar distribution and marker expression (Vogt et al., 2014). To test whether Ctcf-null MGE-derived cells adopt a CGE-like fate, we examined markers of CGE-derived CINs (Vip, Cr, and SP8) (Cai et al., 2013; Ma et al., 2012) in P21 cortex of control and CtcfNkx-Cre animals. No significant difference in either total number (Fig. 7G–L) or laminar position (data not shown) was observed, indicating that Ctcf-null MGE cells, despite expressing low levels of Lhx6, do not ectopically express CGE markers. Together, these data indicate that the Ctcf-null MGE cells that migrate tangentially into the cortex appropriately express Gad1, but do not express Pv or Sst and fail to laminate normally.
Lhx6 reexpression in CTCF-null MGE cells restores SST-expressing interneurons and rescues lamination defects when transplanted in a wild-type host cortex
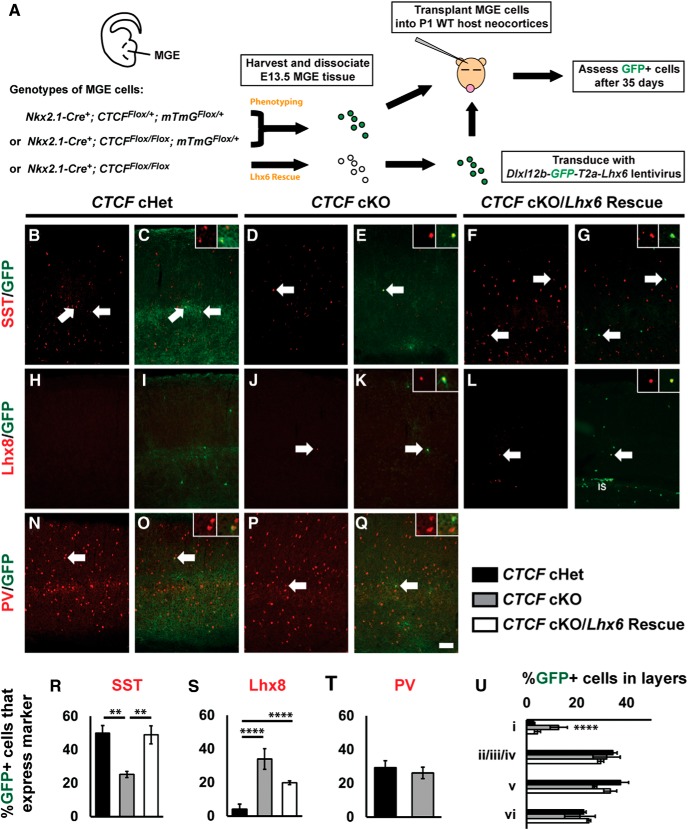
The fate of Ctcf-null MGE interneurons was further evaluated by viral-mediated labeling and transplantation of Ctcf-null or control MGE cells into wild-type host cortex, as described previously (Vogt et al., 2014, 2015). Briefly, the MGE was dissected from E13.5 CtcfNkx-Cre embryos or heterozygous littermate embryos (acting as controls). The MGE was dissociated and either transplanted directly into hosts or transduced with lentiviral constructs before transplantation (Fig. 8A). For phenotyping experiments, the MGE was dissected from CtcfNkx-Cre embryos carrying the Cre-sensitive Rosa-mTmG reporter allele to label cells with GFP. The GFP-labeled MGE cells were then transplanted into P1 wild-type host pups. The brains were removed at P35 and cortical sections were examined by immunofluorescence microscopy.
Figure 8.
Analysis of transplanted CTCF-null MGE cells and rescue by ectopic Lhx6 expression. A, Schema depicting the MGE transplantation and rescue strategy. Briefly, CtcfNkx-Cre heterozygote (cHets) and knock-out (cKO) MGE cells or CTCF cKO MGE cells transduced with a lentivirus expressing Lhx6 were transplanted in the cortex of wild-type neonatal pups. After 35 d, transplanted cells were assessed for GFP and SST (B–G), Lhx8 (H–L), or PV (N–Q). Arrows denote colabeled cells and higher-magnification images of colabeled cells are shown in the top right corners. Quantification of the proportion of GFP+ cells that coexpress SST (R), LHX8 (S), or PV (T). Quantification of the proportion of GFP+ cells that occupy different cortical lamina (U). IS, Injection site. Data are expressed as the mean ± SEM. n = 3 for all groups. **p < 0.01, ****p < 0.0001. Scale bar in Q, 100 μm.
Immunofluorescence staining for SST revealed a marked reduction (∼50%) in the number of transplanted CtcfNkx-Cre GFP+ cells expressing SST compared with controls (Fig. 8B–E,R). Conversely, no changes were observed in PV-expressing cells (Fig. 8N–Q,T). We detected a significantly higher number of GFP+ transplanted cells in layer I (p < 0.0001) and a reduction in cortical layer V, similar to the laminar defects observed in the CtcfNkx-Cre mice at P21 (Fig. 8U). Importantly, a striking increase in the number of transplanted CtcfNkx-Cre GFP+ cells coexpressing Lhx8 was detected (Fig. 8H–K,S). However, these GFP+/ Lhx8+ cells did not migrate into the cortex (Fig. 8H,I), but rather remained close to the injection site (Fig. 9A,B), indicating that their migratory capacity was defective in the cortical environment, as opposed to GFP+/Lhx8− cells.
Figure 9.
Potential alternate fate of CTCF-null MGE cells to GABAergic projection neurons of the basal ganglia. A–H, Transplanted CTCF-null MGE cells remain in proximity to the injection site (IS) and tend to coexpress Lhx8 and GABA, whereas few coexpress ChAT. I, J, A greater number of Lhx8+ and Gbx2+ cells are observed in the basal forebrain of CtcfNes-Cre E15.5 embryonic brain. VZ/SVZ, Ventricular and subventricular zones; FIM, fimbria; at, anterior thalamic nucleus; STT, stria terminalis; gp, globus pallidus.
We reasoned that the reduction of Lhx6 in CTCF-null MGE cells causes the observed phenotypes. To test this, we reexpressed Lhx6 by transducing CtcfNkx-Cre MGE cells (this time not expressing the Rosa-mTmG reporter) with the lentiviral construct (Dlx)I12b-GFP-T2A-Lhx6, as described previously (Vogt et al., 2014) (for schema, see Fig. 8A). At P35, twice as many GFP+ transplanted cells that had been transduced with Lhx6 coexpressed SST (back to normal levels) compared with those transduced with a control vector lacking Lhx6 (Fig. 8R). The lamination defect of CtcfNkx-Cre transplanted cells was also rescued upon reexpression of Lhx6 (Fig. 8U). However, the proportion of GFP+/LHX8+ cells remained high upon forced expression of Lhx6, indicating that Lhx8 upregulation is probably not a compensatory response to reduced Lhx6 levels, but is more likely caused by a failure to repress gene expression in the absence of CTCF. To establish whether the number of transplanted cells was similar between genotypes, we counted the number of GFP+ cells observed for each group and normalized this number to the number of tissue sections used for analysis (Table 2). Although the numbers varied between groups, there were trends toward fewer cells being detected in the CTCF cKO and Lhx6 rescued cells compared with controls, suggesting that some cells might be compromised in the CTCF cKO.
CTCF-null MGE-derived cells may acquire a GABAergic projection neuron fate
Many of the transplanted CTCF-null MGE cells that express LHX8 did not migrate into the cortex and remained close to the injection site (Fig. 9A,B). Given that many LHX8+ cells are fated to become cholinergic striatal neurons, we investigated whether they had gained expression of choline acetyltransferase (ChAT), a marker of cholinergic fate. However, only a small proportion of the transplanted GFP+ cells coexpressed this marker (∼10.5%) and most were located near or within the injection site (Fig. 9C,D,G,H). Fate-mapping studies have shown that ∼60–70% of LHX8+ cells in the basal forebrain become GABAergic instead of cholinergic (Fragkouli et al., 2005), prompting us to examine transplanted cells for coexpression of GABA. The results indicate that the majority of CTCF-null LHX8+ transplanted cells are in fact GABAergic (Fig. 9E,F).
GBX2, a downstream effector of LHX8, marks either striatal cholinergic neurons (Lhx8+, Gbx2+, or ChAT+) or GABAergic projection neurons of the basal forebrain (Lhx8+, Gbx2+, or ChAT−) (Chen et al., 2010). Indeed, upon further examination of the CtcfNes-Cre embryos at E15.5 by ISH, we observed more Gbx2+ and Lhx8+ cells that migrate radially from the MGE to the globus pallidus (Fig. 9E–H), suggesting that loss of Ctcf in MGE cells may respecify a portion of Lhx6+ GABAergic cells destined for the cortex to Lhx8+ GABAergic projection neurons that instead migrate radially into the basal forebrain.
Discussion
The MGE is a transient progenitor zone that generates a variety of different cell types, including GABAergic cortical and striatal interneurons, cholinergic striatal interneurons, and GABAergic and cholinergic projection neurons (Xu et al., 2008; Fragkouli et al., 2009; Flandin et al., 2010; Zechel et al., 2014). The mechanism of genetic programming in the MGE that allows for this cellular diversity is the subject of intense research. The present study advances knowledge of these events by demonstrating that CTCF, a factor involved in higher-order organization of chromatin structure, is required for fate specification of interneurons born in the MGE. Loss of CTCF alters the balance between GABAergic cells fated to become CINs versus basal forebrain projection neurons through the regulation of the LHX6 and LHX8 LIM homeodomain factors.
Expression of Lhx6 and Lhx8 was altered upon Ctcf inactivation, whereas the upstream regulator NKX2.1 remained unaffected. Moreover, we found no evidence that CTCF interacts with NKX2.1 or cooccupies binding sites in E13.5 MGE chromatin around Lhx6 and Lhx8 gene loci. Alternate possibilities for regulation must therefore be considered, such as effects on another NKX2.1-binding partner upon Ctcf deletion or that CTCF is required to set up the specific chromatin architecture permissive for NKX2.1 binding without direct protein–protein interaction.
Deletion of another genome organizer, Special AT-rich DNA Binding Protein 1 (Satb1), also impairs differentiation of SST+ interneurons (Denaxa et al., 2012), corroborating the idea that chromatin organization is a key regulatory mechanism of cell fate specification and differentiation of MGE-derived cells. CTCF has been shown in other models to influence gene expression by modulating looping, topological chromatin domains, and enhancer binding (Oti et al., 2016). Additional studies will be required to precisely uncover how CTCF affects chromatin structure and gene expression in the MGE.
Decreased expression of Lhx6 upon loss of Ctcf is associated with fewer Sst+ and Pv+ interneurons in the cortex. We were able to show that the effect on Sst+ interneurons results from low Lhx6 expression because lentiviral transduction of Lhx6 in Ctcf-null MGE cells fully reinstated the number of Sst+ CINs to control levels in transplantation experiments. However, the transplantation studies failed to replicate a loss of PV+ interneurons seen in the CtcfNkx-Cre model. Transplantation experiments were performed in a CD1 background to maximize litter size, whereas the P20–P22 CtcfNkx-Cre mice were generated in the C57BL/6 strain. It is therefore conceivable that there is a variable necessity for Lhx6 expression in different mouse strains/genetic backgrounds for Pv expression. Alternatively, the Pv+ interneurons that are lost in the genetic mouse model may fail to reach the neocortex. Although transplanted MGE cells can still migrate to the neocortex where they are injected, they do not experience the same obstacles encountered by endogenous MGE-derived cells that migrate tangentially into the neocortex. It is possible that the cells fated to become Pv+ may be more affected by migration than by fate, although this remains to be determined. Additionally, a similar phenomenon has been described previously (Neves et al., 2013) using a hypomorphic allele of Lhx6, which resulted in loss of SST, but not PV interneuron subtypes. Together with the work presented here, these data suggest that the role of LHX6 is stricter for Sst+ subtype specification and that its role in Pv+ subtype specification may be compensated for in different genetic backgrounds.
Lhx6-null MGE cells were previously found to undergo a partial fate switch to CGE-like cells (Vogt et al., 2014), but we did not observe this phenomenon in the CTCF-null MGE cells, indicating that only complete loss of Lhx6 expression enacts a switch to CGE-like fate. Lhx6 deletion models show defective tangential migration of interneurons into the cortex, which we also observed with deletion of Ctcf in the MGE. Reduced expression of the CXCR4 chemoreceptor would be predicted to contribute to this phenotype. By weaning age, however, the Ctcf mutants had the same number of Gad1+ CINs, suggesting that misspecified CTCF-null MGE cells have delayed tangential migration but eventually populate the cortex, albeit with abnormal distribution, based on Reelin positivity. These findings also suggest that CTCF-null CINs are appropriately specified to GABAergic fate but fail to express Pv or Sst. This is not surprising because the factors that govern Gad1 expression are controlled by different transcriptional pathways that work in parallel to LHX6/LHX8. Lhx6 mutants remain GAD1+ but exhibit an MGE-to-CGE fate switch (Liodis et al., 2007; Vogt et al., 2014) and Nkx2.1 mutants remain GAD1+ with an MGE-to-LGE fate switch (Sussel et al., 1999).
Ctcf deletion in MGE cells also resulted in increased expression of Lhx8, which is an interesting observation because it could explain the higher number of Gbx2+ cells in the basal forebrain in the CtcfNes-Cre embryos. These cells could potentially represent basal forebrain GABAergic neurons that project to the cortex. This is supported by the observation that transplanted Ctcf-null cells coexpressed LHX8 and GABA at the injection site, whereas very few expressed ChAT, a marker of cholinergic neurons. Reexpression of Lhx6 was not sufficient to reduce the number of LHX8+ cells to normal levels in transplantations, which could be explained by reduced functionality of the DlxI12b enhancer to reexpress Lhx6 in Lhx8+ Ctcf-null cells or may suggest that LHX8 overexpression is a result of CTCF loss and not downstream of reduced Lhx6 expression.
Understanding the molecular control switches in cell fate specification of MGE cells is relevant to the in vitro differentiation of these progenitors to specific neuron types and in the treatment of human neurological disorders (Danjo et al., 2011; Chohan and Moore, 2016; Chen et al., 2016). The genetic pathways in MGE differentiation and specification are highly conserved between mouse and human (Hansen et al., 2013; Wang et al., 2014; Ahn et al., 2016), so delineating the molecular events that regulate MGE development can improve our ability to effectively generate MGE-derived cell subtypes from human stem cells. CTCF mutations in humans can result in intellectual disability (Gregor et al., 2013), ASD (Chang et al., 2015), and conditions with altered GABAergic neuron function (Blatt and Fatemi, 2011; Smith-Hicks, 2013). In addition, CTCF has been associated with schizophrenia (Juraeva et al., 2014), which has also been linked with defects in LHX6 (Volk et al., 2014). Our study implicates CTCF in fate specification and migration of MGE progenitors and complements current efforts to understand MGE development from the perspective of chromatin regulators.
Footnotes
This work was supported by the Canadian Institutes for Health Research (Grant MOP142268 to N.G.B.) and the National Institutes of Mental Health–National Institutes of Health (Grant R01 MH081880 to J.R.). A.E. was supported by scholarship funding from CIHR Vanier and Ontario Mental Health Foundation. A.W. was supported by a Natural Science and Engineering Research Council (NSERC) Canadian Graduate Scholarship. We thank Dr. Neils Galjart for the Ctcf floxed mice and Dr. Aleksandar Rajkovic for the Lhx8 antibody.
J.R. is cofounder, stockholder, and currently on the scientific board of Neurona, a company studying the potential therapeutic use of interneuron transplantation. The remaining authors declare no competing financial interests.
References
- Abe P, Molnár Z, Tzeng YS, Lai DM, Arnold SJ, Stumm R (2015) Intermediate progenitors facilitate intracortical progression of thalamocortical axons and interneurons through CXCL12 chemokine signaling. J Neurosci 35:13053–13063. 10.1523/JNEUROSCI.1488-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S, Kim TG, Kim KS, Chung S (2016) Differentiation of human pluripotent stem cells into medial ganglionic eminence vs. caudal ganglionic eminence cells. Methods 101:103–112. 10.1016/j.ymeth.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastaki F, Nair P, Mohamed M, Malik EM, Helmi M, Al-Ali MT, Hamzeh AR (2017) Identification of a novel CTCF mutation responsible for syndromic intellectual disability: a case report. BMC Med Genet 18:68. 10.1186/s12881-017-0429-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérubé NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ (2005) The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest 115:258–267. 10.1172/JCI200522329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt GJ, Fatemi SH (2011) Alterations in GABAergic biomarkers in the autism brain: research findings and clinical implications. Anat Rec (Hoboken) 294:1646–1652. 10.1002/ar.21252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Zhang Q, Wang C, Zhang Y, Ma T, Zhou X, Tian M, Rubenstein JL, Yang Z (2013) Nuclear receptor COUP-TFII-expressing neocortical interneurons are derived from the medial and lateral/caudal ganglionic eminence and define specific subsets of mature interneurons. J Comp Neurol 521:479–497. 10.1002/cne.23186 [DOI] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D (2015) Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci 18:191–198. 10.1038/nn.3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Plocik A, Anderson NC, Moakley D, Boyi T, Dundes C, Lassiter C, Graveley BR, Grabel L (2016) Transcriptome and in vitro differentiation profile of human embryonic stem cell derived NKX2.1-positive neural progenitors. Stem Cell Rev 12:744–756. 10.1007/s12015-016-9676-2 [DOI] [PubMed] [Google Scholar]
- Chen L, Chatterjee M, Li JY (2010) The mouse homeobox gene Gbx2 is required for the development of cholinergic interneurons in the striatum. J Neurosci 30:14824–14834. 10.1523/JNEUROSCI.3742-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Vogt D, Wang Y, Visel A, Silberberg SN, Nicholas CR, Danjo T, Pollack JL, Pennacchio LA, Anderson S, Sasai Y, Baraban SC, Kriegstein AR, Alvarez-Buylla A, Rubenstein JL (2013) Use of “MGE enhancers” for labeling and selection of embryonic stem cell-derived medial ganglionic eminence (MGE) progenitors and neurons. PLoS One 8:e61956. 10.1371/journal.pone.0061956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chohan MO, Moore H (2016) Interneuron progenitor transplantation to treat CNS dysfunction. Front Neural Circuits 10:64. 10.3389/fncir.2016.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danjo T, Eiraku M, Muguruma K, Watanabe K, Kawada M, Yanagawa Y, Rubenstein JL, Sasai Y (2011) Subregional specification of embryonic stem cell-derived ventral telencephalic tissues by timed and combinatory treatment with extrinsic signals. J Neurosci 31:1919–1933. 10.1523/JNEUROSCI.5128-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio JA, Soriano E, Ferrer I (1992) Development of GABA-immunoreactivity in the neocortex of the mouse. J Comp Neurol 326:501–526. 10.1002/cne.903260403 [DOI] [PubMed] [Google Scholar]
- Denaxa M, Kalaitzidou M, Garefalaki A, Achimastou A, Lasrado R, Maes T, Pachnis V (2012) Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Reports 2:1351–1362. 10.1016/j.celrep.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-Seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Xu Q, Ocbina PJ, Anderson SA (2008) NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development 135:1559–1567. 10.1242/dev.015123 [DOI] [PubMed] [Google Scholar]
- Flandin P, Kimura S, Rubenstein JL (2010) The progenitor zone of the ventral medial ganglionic eminence requires Nkx2–1 to generate most of the globus pallidus but few neocortical interneurons. J Neurosci 30:2812–2823. 10.1523/JNEUROSCI.4228-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, Hearn C, Errington M, Cooke S, Grigoriou M, Bliss T, Stylianopoulou F, Pachnis V (2005) Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. Eur J Neurosci 21:2923–2938. 10.1111/j.1460-9568.2005.04141.x [DOI] [PubMed] [Google Scholar]
- Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V (2009) LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development 136:3841–3851. 10.1242/dev.038083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JR, Bartley AF, Hays SA, Huber KM (2008) Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J Neurophysiol 100:2615–2626. 10.1152/jn.90752.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Espinosa JM (2010) Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-cohesin binding. Genes Dev 24:1022–1034. 10.1101/gad.1881010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, Kjaergaard S, Rauch A, Stunnenberg HG, Uebe S, Vasileiou G, Reis A, Zhou H, Zweier C (2013) De novo mutations in the genome organizer CTCF cause intellectual disability. Am J Hum Genet 93:124–131. 10.1016/j.ajhg.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad M, Schmidt SL, Zhang X, Bray R, Frohlich F, Ghashghaei HT (2015) Transplantation of GABAergic interneurons into the neonatal primary visual cortex reduces absence seizures in stargazer mice. Cereb Cortex 25:2970–2979. 10.1093/cercor/bhu094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, Alvarez-Buylla A, Kriegstein AR (2013) Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci 16:1576–1587. 10.1038/nn.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi E, Ariza J, Rogers H, Noctor SC, Martínez-Cerdeño V (2017) The number of parvalbumin-expressing interneurons is decreased in the medial prefrontal cortex in autism. Cereb Cortex 27:1931–1943. 10.1093/cercor/bhw021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath H, Ribeiro de Almeida C, de Sleutels F, Dingjan G, van de Nobelen S, Jonkers I, Ling KW, Gribnau J, Renkawitz R, Grosveld F, Hendriks RW, Galjart N (2008) CTCF regulates cell cycle progression of αβ T cells in the thymus. EMBO J 27:2839–2850. 10.1038/emboj.2008.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T (2012) CTCF is required for neural development and stochastic expression of clustered pcdh genes in neurons. Cell Reports 2:345–357. 10.1016/j.celrep.2012.06.014 [DOI] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, et al. (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature 515:216–221. 10.1038/nature13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juraeva D, Haenisch B, Zapatka M, Frank J, GROUP Investigators, PSYCH-GEMS SCZ Working Group, Witt SH, Mühleisen TW, Treutlein J, Strohmaier J, Meier S, Degenhardt F, Giegling I, Ripke S, Leber M, Lange C, Schulze TG, Mössner R, Nenadic I, Sauer H, et al. (2014) Integrated pathway-based approach identifies association between genomic regions at CTCF and CACNB2 and schizophrenia. PLoS Genet 10:e1004345. 10.1371/journal.pgen.1004345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ (2013) Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V (2007) Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci 27:3078–3089. 10.1523/JNEUROSCI.3055-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Martinez-Cerdeno V, Hagerman RJ (2015) Advances in the understanding of the gabaergic neurobiology of FMR1 expanded alleles leading to targeted treatments for fragile X spectrum disorder. Curr Pharm Des 21:4972–4979. 10.2174/1381612821666150914121038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Zhang Q, Cai Y, You Y, Rubenstein JL, Yang Z (2012) A subpopulation of dorsal Lateral/Caudal ganglionic eminence-derived neocortical interneurons expresses the transcription factor Sp8. Cereb Cortex 22:2120–2130. 10.1093/cercor/bhr296 [DOI] [PubMed] [Google Scholar]
- Marín O. (2013) Cellular and molecular mechanisms controlling the migration of neocortical interneurons. Eur J Neurosci 38:2019–2029. 10.1111/ejn.12225 [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL (2000) Origin and molecular specification of striatal interneurons. J Neurosci 20:6063–6076. 10.1523/JNEUROSCI.20-16-06063.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Nora EP (2016) CTCF and cohesin in genome folding and transcriptional gene regulation. Annu Rev Genomics Hum Genet 17:17–43. 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- Minoo P, Su G, Drum H, Bringas P, Kimura S (1999) Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(-/-) mouse embryos. Dev Biol 209:60–71. 10.1006/dbio.1999.9234 [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Fishell G (2011) GABAergic interneuron lineages selectively sort into specific cortical layers during early postnatal development. Cereb Cortex 21:845–852. 10.1093/cercor/bhq155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Shah MM, Liodis P, Achimastou A, Denaxa M, Roalfe G, Sesay A, Walker MC, Pachnis V (2013) The LIM homeodomain protein Lhx6 regulates maturation of interneurons and network excitability in the mammalian cortex. Cereb Cortex 23:1811–1823. 10.1093/cercor/bhs159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oti M, Falck J, Huynen MA, Zhou H (2016) CTCF-mediated chromatin loops enclose inducible gene regulatory domains. BMC Genomics 17:252. 10.1186/s12864-016-2516-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluszkiewicz SM, Olmos-Serrano JL, Corbin JG, Huntsman MM (2011) Impaired inhibitory control of cortical synchronization in fragile X syndrome. J Neurophysiol 106:2264–2272. 10.1152/jn.00421.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cremades D, Hernández S, Blasco-Ibáñez JM, Crespo C, Nacher J, Varea E (2010) Alteration of inhibitory circuits in the somatosensory cortex of Ts65Dn mice, a model for Down's syndrome. J Neural Transm (Vienna) 117:445–455. 10.1007/s00702-010-0376-9 [DOI] [PubMed] [Google Scholar]
- Potier MC, Braudeau J, Dauphinot L, Delatour B (2014) Reducing GABAergic inhibition restores cognitive functions in a mouse model of down syndrome. CNS Neurol Disord Drug Targets 13:8–15. 10.2174/18715273113126660185 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford) 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T (2014) deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42(Web Server issue):W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Mejias M, Martinez de Lagran M, Mattia M, Castano-Prat P, Perez-Mendez L, Ciria-Suarez L, Gener T, Sancristobal B, García-Ojalvo J, Gruart A, Delgado-García JM, Sanchez-Vives MV, Dierssen M (2016) Overexpression of Dyrk1A, a down syndrome candidate, decreases excitability and impairs gamma oscillations in the prefrontal cortex. J Neurosci 36:3648–3659. 10.1523/JNEUROSCI.2517-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sams DS, Nardone S, Getselter D, Raz D, Tal M, Rayi PR, Kaphzan H, Hakim O, Elliott E (2016) Neuronal CTCF is necessary for basal and experience-dependent gene regulation, memory formation, and genomic structure of BDNF and Arc. Cell Reports 17:2418–2430. 10.1016/j.celrep.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Sandberg M, Flandin P, Silberberg S, Su-Feher L, Price JD, Hu JS, Kim C, Visel A, Nord AS, Rubenstein JLR (2016) Transcriptional networks controlled by NKX2–1 in the development of forebrain GABAergic neurons. Neuron 91:1260–1275. 10.1016/j.neuron.2016.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Hicks CL. (2013) GABAergic dysfunction in pediatric neuro-developmental disorders. Front Cell Neurosci 7:269. 10.3389/fncel.2013.00269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell DG, Nicholas CR, Basbaum AI, Stryker MP, Kriegstein AR, Rubenstein JL, Alvarez-Buylla A (2014) Interneurons from embryonic development to cell-based therapy. Science 344:1240622. 10.1126/science.1240622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL (1999) Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126:3359–3370. [DOI] [PubMed] [Google Scholar]
- Tremblay R, Lee S, Rudy B (2016) GABAergic interneurons in the neocortex: from cellular properties to circuits. Neuron 91:260–292. 10.1016/j.neuron.2016.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt D, Hunt RF, Mandal S, Sandberg M, Silberberg SN, Nagasawa T, Yang Z, Baraban SC, Rubenstein JL (2014) Lhx6 directly regulates arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron 82:350–364. 10.1016/j.neuron.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt D, Wu PR, Sorrells SF, Arnold C, Alvarez-Buylla A, Rubenstein JLR (2015) Viral-mediated labeling and transplantation of medial ganglionic eminence (MGE) cells for in vivo studies. J Vis Exp 98. 10.3791/52740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Edelson JR, Lewis DA (2014) Cortical inhibitory neuron disturbances in schizophrenia: role of the ontogenetic transcription factor Lhx6. Schizophr Bull 40:1053–1061. 10.1093/schbul/sbu068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, You Y, Qi D, Zhou X, Wang L, Wei S, Zhang Z, Huang W, Liu Z, Liu F, Ma L, Yang Z (2014) Human and monkey striatal interneurons are derived from the medial ganglionic eminence but not from the adult subventricular zone. J Neurosci 34:10906–10923. 10.1523/JNEUROSCI.1758-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson LA, Wang X, Elbert A, Kernohan KD, Galjart N, Bérubé NG (2014) Dual effect of CTCF loss on neuroprogenitor differentiation and survival. J Neurosci 34:2860–2870. 10.1523/JNEUROSCI.3769-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA (2004) Origins of cortical interneuron subtypes. J Neurosci 24:2612–2622. 10.1523/JNEUROSCI.5667-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA (2008) Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol 506:16–29. 10.1002/cne.21529 [DOI] [PubMed] [Google Scholar]
- Zechel S, Zajac P, Lönnerberg P, Ibáñez CF, Linnarsson S (2014) Topographical transcriptome mapping of the mouse medial ganglionic eminence by spatially resolved RNA-Seq. Genome Biol 15:486. 10.1186/s13059-014-0486-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-seq (MACS). Genome Biol 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Marín O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JL, Westphal H (2003) The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci U S A 100:9005–9010. 10.1073/pnas.1537759100 [DOI] [PMC free article] [PubMed] [Google Scholar]