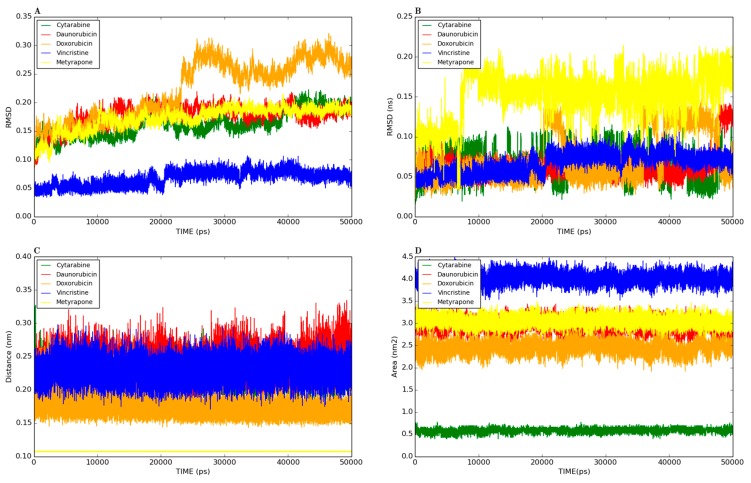
Figure 2.
Analyses of the productive binding poses of cytarabine, daunorubicin, doxorubicin, vincristine, cytochrome (alone) and metyrapone (control) were subjected to 50-ns simulations. (A) Root mean square deviation (RMSD) of the protein backbone atoms, with respect to the initial structure; (B) drug RMSD, with respect to the initial structure; (C) The solvent accessible surface area for the drug complexes; (D) heme to drug distances measured.