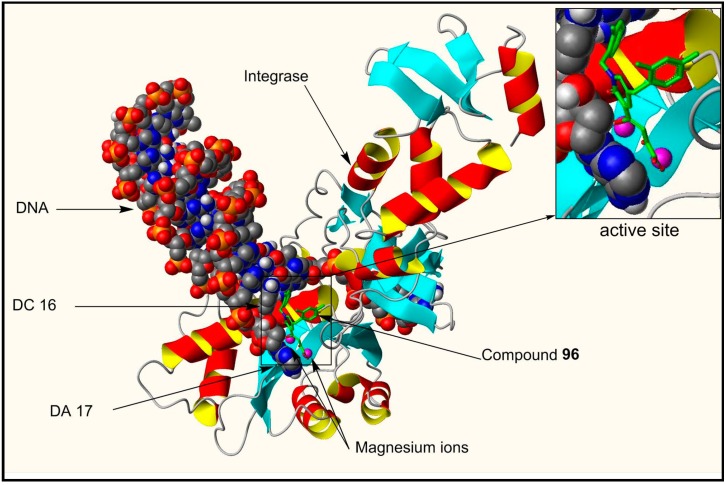
Figure 7.
Docking pose of compound 96 (stick model in green) within the active site of modeled HIV-1integrase intasome. The inset illustrates the chelation of the two magnesium metals (indicated as purple spheres) with the inhibitor. The docking picture also shows ring-stacking interactions of compound 96 with DC 16 (deoxycytidine 16) as well as with DA 17 (deoxyadenosine 17) within the active site. Some of the key amino acid residues in integrase interacting with compound 96 are as follows: A/Asp64, A/Asp116, A/Tyr143, A/Gln144, A/Pro145, A/Gln148, A/Gly149, and A/Glu152. Molecular modeling of the crystal structure of prototype foamy virus (PFV) integrase intasome (PDB code 3OYA) [15,60] with compound 96 docked within the catalytic site was achieved by using the Surflex-Dock package within Sybyl-X [Sybyl-X1.3 (winnt_os5x) version] (Tripos, St. Louis, MO, USA, 2011).