Figure 2.
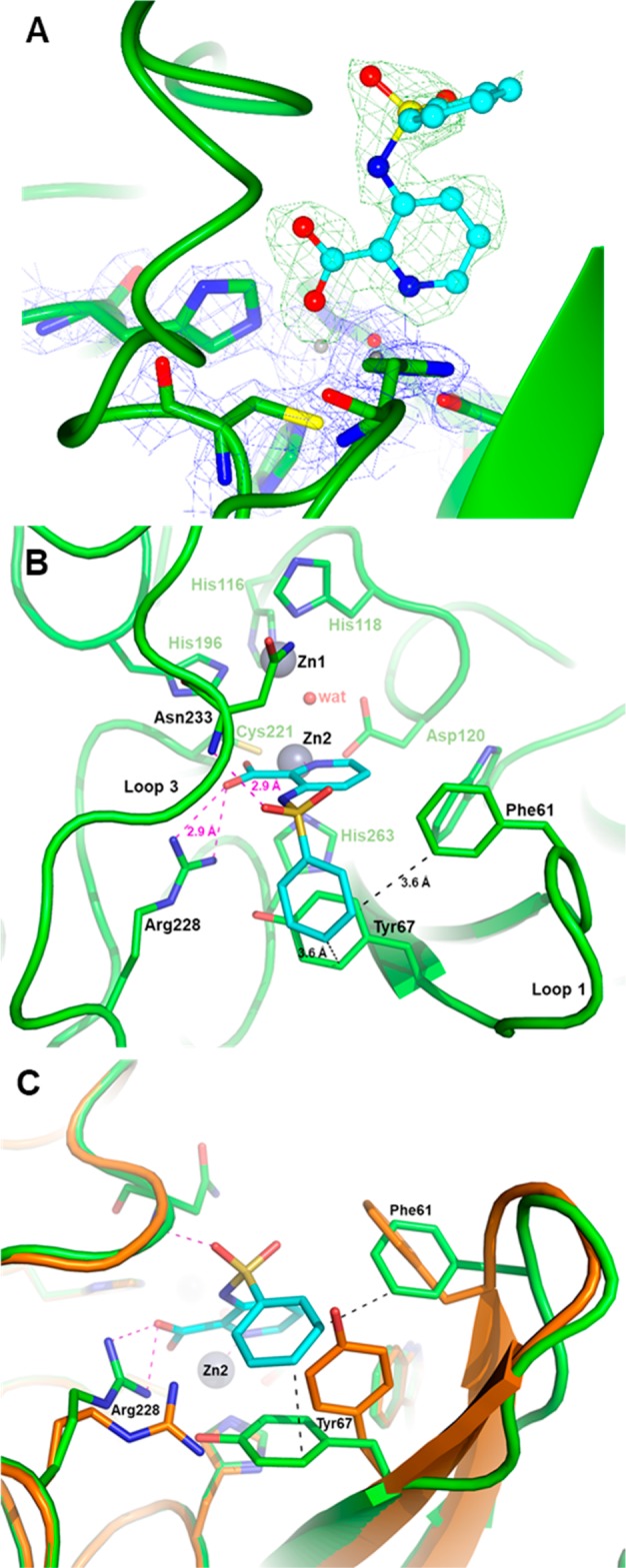
X-ray structure of VIM-2 inhibited by 32. (A) Close view of the VIM-2 active site (protein secondary structure elements and the active-site residues are shown in green, surrounded by the 2Fo–Fc Fourier map, blue meshes, contoured at 2 σ) in complex with compound 32 (shown in cyan and surrounded by the omit Fo–Fc map, green meshes, contoured at 3 σ). (B) Close-up view of the VIM-2 active site showing the network of interactions between the inhibitor (cyan), the Zn2 ion and residues Arg228 and Asn233. (C) Orthogonal view of panel B and superimposition with the VIM-2 native structure (PDB code 1KO3, orange) showing the significant movements of the side chains of residues Phe61, Tyr67, and Arg228 (green sticks) upon the binding of 32 (cyan).
