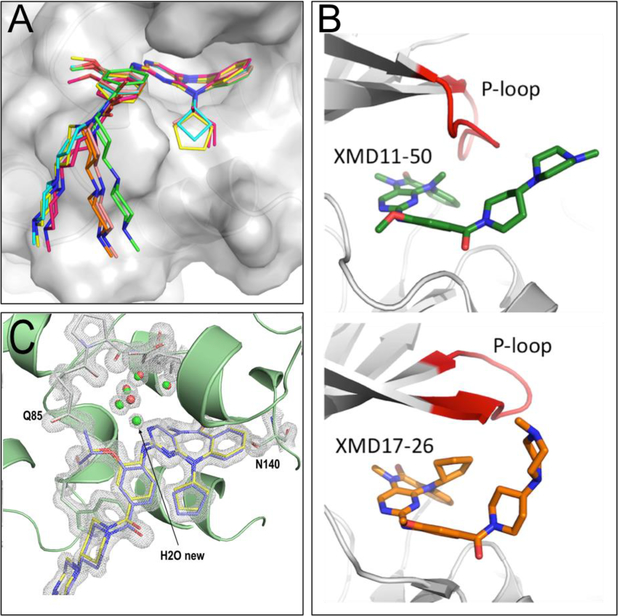
Figure 3.
(A) Overlay of structures of XMD11–50 (green), JWG-048 (salmon), JWG-046 (orange), JWG-071(pink), JWG-069 (cyan) and XMD17–26 (yellow) highlighting the different curvature of the diazipinone ring system and deviation of the tail. (B) Overlay of Roco4/XMD11–50 (pdb code 4YZM, top), and ERK5/XMD17–26 (pdb code 4B99, bottom), crystal structure reveal differences in P-loop (red) conformation and explain observed R2 SAR. (C) Overlay of JWG-047 and XMD17–26 in complex with BRD4-BD1 reveal differences in local interactions and the presence of a new water molecule in the JWG-047. Composite-omit map of JWG-047 in 2Fo-Fc format with a σ-level at 2.0.