Table 2.
Activity profile of inhibitors with diazepinone scaffold
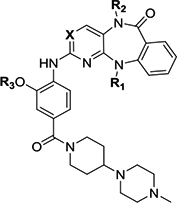 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound ID | R1 | R2 | R3 | X | Biochemical IC50 (BRD4, μM)a | Biochemical IC50 (BRD4, μM)b | Enzymatic IC50 (ERK5, μM)c | Enzymatic IC50 (LRRK2, μM)d | Viability IC50(BRD4, μM)e | Cellular EC50 (ERK5, μM)f | Inhibitor classificationg | Co-crystal (pdb number) |
| XMD11–50 (LRRK2-IN-1) | Me | Me | Me | N | 0.703 ± 0.017 | 1.04 ± 0.18 | 0.114 ±0.011 | 0.0034 ±0.0017 | 1.989 ± 0.299 | 0.16 ± 0.04 | BRD4/LRRK2 | Roco4(4YZM)32 BRD4 (5WA5) |
| JWG-048 | Et | Me | Me | N | 1.33 ± 0.05 | 2.41 ± 0.23 | 0.173 ±0.037 | 0.0044 ±0.0005 | 2.625 ± 0.540 | 0.022 ± 0.004 | LRRK2 | BRD4 (5W55) |
| JWG-046 | i-Pr | Me | Me | N | 4.59 ± 0.14 | 5.14 ± 0.59 | 0.125 ±0.026 | 0.054 ± 0.004 | ND | 0.014 ± 0.004 | ERK5/LRRK2 | BRD4 (6CD4) |
| JWG-071 | s-Bu | Me | Me | N | 5.42 ± 0.18 | 6.31 ± 1.93 | 0.088 ±0.005 | 0.109 ± 0.014 | >10 | 0.020 ± 0.003 | ERK5/LRRK2 | BRD4 (6CJ1) |
| JWG-069 | cyclobutyl | Me | Me | N | 0.201 ± 0.007 | 0.41 ± 0.01 | 0.075 ±0.009 | 0.021 ± 0.001 | 0.116 ± 0.001 | 0.019 ± 0.005 | BRD4/ERK5/LRRK2 | BRD4 (6CIY) |
| XMD17–26 | cyclopentyl | Me | Me | N | 0.760 ± 0.012 | 0.75 ± 0.11 | 0.082 ±0.009 | 0.095 ± 0.010 | 0.664 ± 0.014 | 0.08 ± 0.02 | BRD4/ERK5/LRRK2 | ERK5 (4B99)21 BRD4 (5CD5) |
| JWG-112 | s-Bu | H | Me | N | 25.7 ± 1.8 | >40 | >10 | 1.59 ± 0.37 | ND | ND | No activity | |
| JWG-049 | i-Pr | Me | Et | N | 2.50 ± 0.07 | 3.53 ± 0.82 | 0.069 ±0.007 | 0.114 ± 0.011 | ND | 0.025 ± 0.007 | ERK5/LRRK2 | |
| JWG-114 | i-Pr | Me | i-Pr | N | 0.862 ± 0.065 | 1.08 ± 0.13 | 0.421 ±0.007 | 0.235 ± 0.034 | ND | ND | BRD4/weak LRRK2 | |
| XMD17–109 (ERK5-IN-1) | cyclopentyl | Me | Et | N | 0.217 ± 0.008 | 0.68 ± 0.03 | 0.162 ±0.006 | 0.171 ± 0.030 | ND | 0.09 ± 0.03 | BRD4/ERK5/LRRK2 | |
| JWG-047 | cyclopentyl | Me | i-Pr | N | 0.135 ± 0.004 | 0.30 ± 0.05 | 0.160 ±0.008 | 0.296 ± 0.031 | 0.465 ± 0.012 | 0.032 ± 0.009 | BRD4/ERK5/LRRK2 | BRD4 (6CIS) |
| AX15836 | SO2Me | Me | Et | N | >100 | >40 | 0.012 ±0.002 | 0.942 ± 0.569 | ND | 0.036 ± 0.004 | ERK5 | |
| DFCI-2–208 | Me | Me | Me | C | 5.21 ± 0.14 | 4.64 ± 0.65 | >10 | >10 | ND | ND | No activity | |
| JWG-115 | cyclopentyl | Me | i-Pr | C | 1.08 ± 0.05 | 1.10 ± 0.05 | >10 | > 3.3 | ND | ND | BRD4 (weak) | |
| (+)-JQ-1 | 0.064 | 0.12 | >10 | >10 | 0.228 | ND | BRD4 | |||||
IC50 values were measured by AlphaScreen™ binding assay and reported as the average of 2 replicates ± SE. Experiments were conducted in Dana-Farber Cancer Institute.
IC50 values were measured by AlphaScreen™ binding assay and reported as the average of 4 replicates ± SD. Experiments were conducted in University of Oxford.
IC50 values were measured by in vitro assay and reported as the average of 2 replicates ± SD.
IC50 values were measured using Adapta assay format (ThermoFisher Scientific) and reported as the average of 2 replicates ± SD.
IC50 were determined in 797 NUT-midline carcinoma cells by cell numbers using Cell Titer Glo and reported as the average of 3 replicates ± SD.
EC50 values were measured by EGF-stimulated autophosphorylation of ERK5 in Hela cells and reported as the average of 2 replicates ± SD.
Considering that weak BET bromodomain inhibition has been shown to result in phenotypic responses, a BRD4 activity within 20 fold of JQ1 was defined as BET active. 0.3 μM was chosen as the threshold for kinase activity. A 10-fold activity difference among kinases was considered selective.
