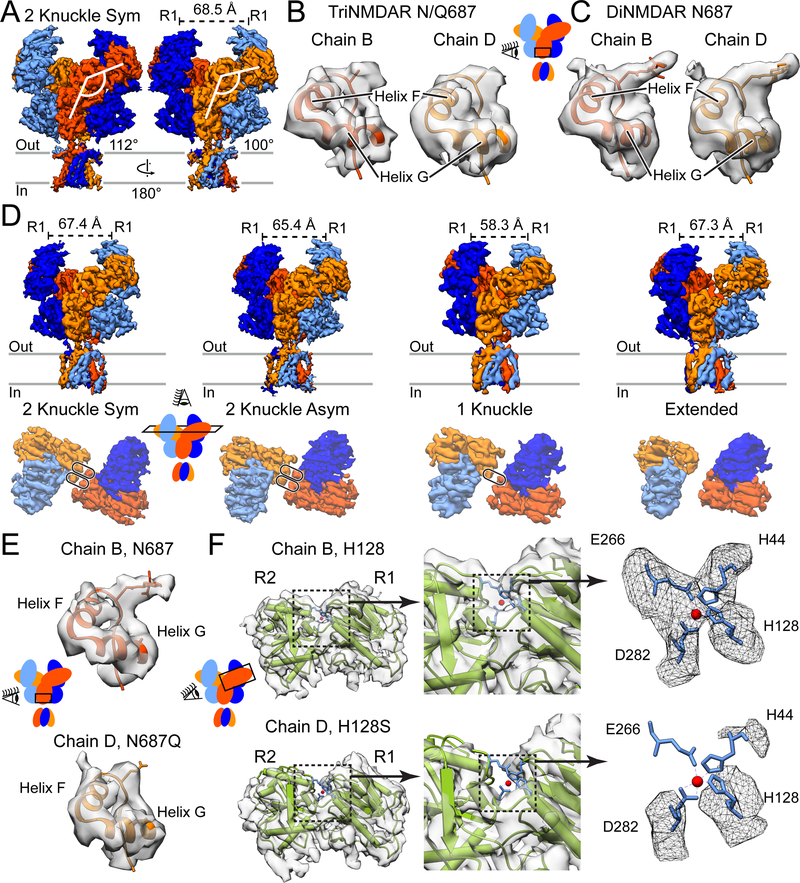
Figure 3. CryoEM Structures of the GluN1/GluN2A/GluN2A* triNMDAR.
(A) Side views of the GluN1/GluN2A/GluN2A* triNMDAR cryoEM map in the presence of 1 mM glutamate and glycine at pH 7.4 with 1 mM EDTA. The distance between COM of the upper lobes (R1) of GluN1 ATD, as well as the ATD-LBD angle for the two GluN2A subunits, are shown. See also Figure S2.
(B and C) Atomic model and the associated cryoEM density for the GluN2 subunit of the triNMDAR (B) and the diNMDAR (C) in the vicinity of residue 687 in the LBD D2 lobe. There is density consistent with glycosylation of N687 in the diNMDAR, however, this density is not present in either subunit for the triNMDAR. Since distinguishing between GluN2A and GluN2A* subunits is not possible based on glycosylation at residue 687, residue 687 in the map is represented as an alanine.
(D) Side view of the full receptor (top) and top view of the ATD (bottom) of the GluN1/GluN2A/GluN2A* triNMDAR cryoEM map in the presence of 1 mM glutamate and glycine and 1 μM zinc at pH 7.4. See also Figures S1 and S5.
(E) Atomic model and the associated map for the GluN2A subunit (Chain B, top) and the GluN2A* subunit (Chain D, bottom) in the vicinity of residue 687 for the triNMDAR 2-knuckle-asym conformation in the presence of 1 μM zinc. At the same contour level, there is density consistent with glycosylation at only one GluN2 subunit, allowing for the identification of the wt GluN2A and the H128S & N687Q mutant GluN2A* subunits.
(F) The crystal structure of the GluN2A ATD bound by zinc (PDB: 5TPW) fitted into the cryoEM map of the two GluN2 subunits of the triNMDAR 2-knuckle-asym conformation with the R1 lobes aligned. There is density consistent with four amino acid sidechains (H44, H128, E266, D282) coordinating zinc for the WT-subunit. At the same contour level, there is no density in the vicinity of the zinc-binding pocket for the GluN2A* subunit with the H128S mutation.