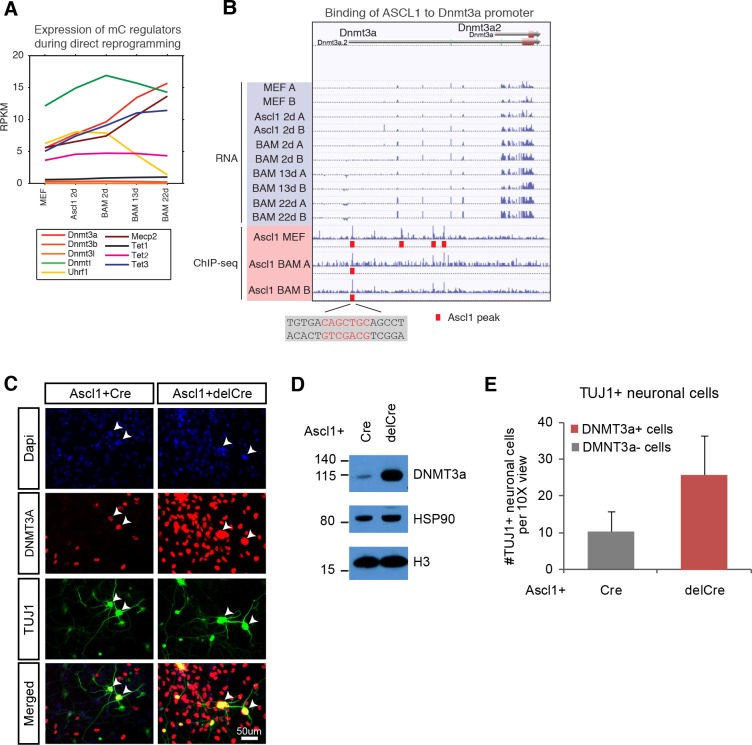
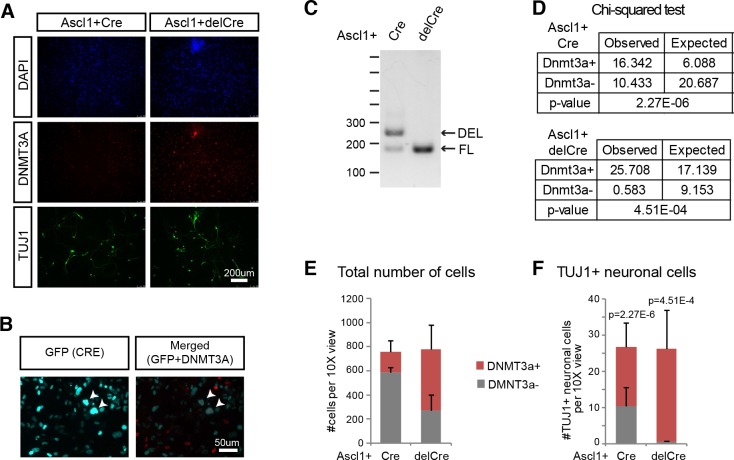
Figure 5. de novo DNA methylation is required for efficient reprogramming from fibroblasts to neurons.
(A) Expression of DNA methylation regulators and readers during neuronal reprogramming. (B) Ascl1 ChIP-seq peak is located proximal to Dnmt3a promoter. (C) Immunostaining of Dnmt3a/3bfl/fl cells 14d post-Ascl1 induction with dox. There appears to be a similar reprogramming efficiency when comparing the CreGfp (left) and delCreGfp (right) conditions. However, most TUJ1 +iN cells in the CreGfp condition were still expressing low levels of DNMT3A (white arrows) despite co-infection with CreGfp. (D) Western blot showing efficient knock-out of DNMT3A in Ascl1 +Cre expressing cells compared to Ascl1 +delCre control 13 days post induction of Ascl1. (E) Average counts of TUJ1 +iN cells per 10X field of view 14d post-Ascl1 induction that are co-infected with 1) CreGfp and Dnmt3a- (gray) or 2) delCreGfp and Dnmt3a+ (red) (error bars are stderr, n = 3).