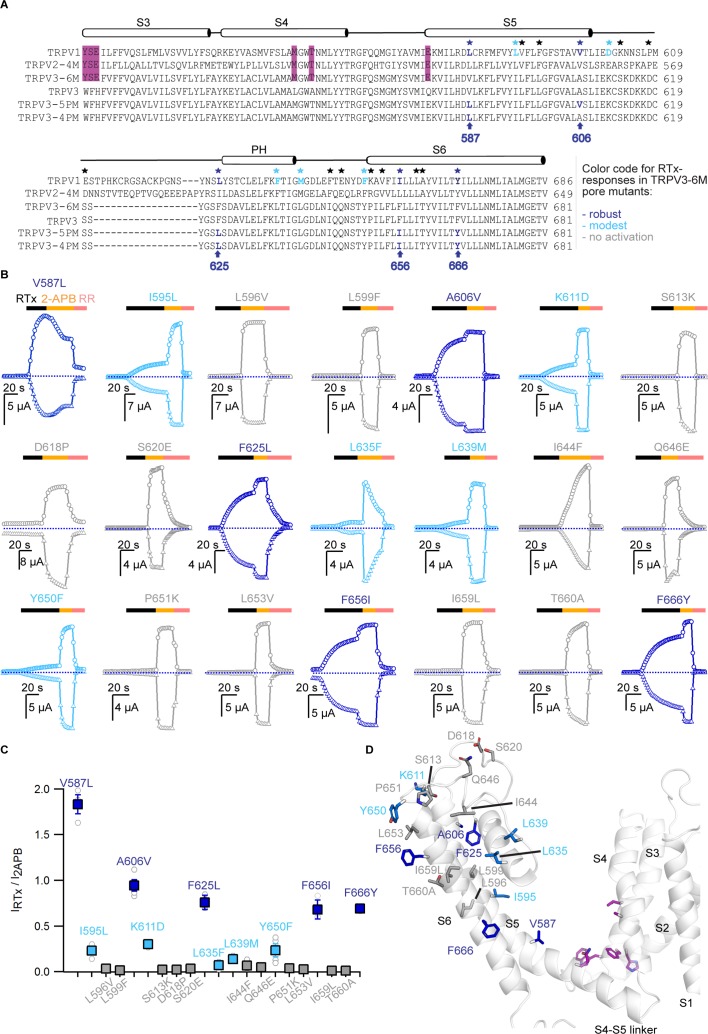
Figure 2. Identification of key residues for RTx activation in the pore domain of TRPV3-6M.
(A) Sequence alignment of the S3 to S6 TM helices for rTRPV1, rTRPV2-4M, mTRPV3-6M, WT mTRPV3, TRPV3-5PM and TRPV3-4PM. Residues labeled with stars are conserved in TRPV1 and TRPV2 but different in TRPV3; black stars – mutations that did not influence RTx activation; blue - mutations that enabled moderate (light blue) or strong (dark blue) responses to RTx. The purple highlights denote the 6M mutations. (B) Representative time courses of activation of TRPV3-6M channels with individual pore mutations. Channels were stimulated by RTx (100 nM) and 2-APB (3 mM), and blocked with RR (50 μM) as indicated by the colored horizontal lines. Currents were measured at +60 (circles) and −60 mV (triangles) as in Figure 1. The dotted horizontal lines indicate the zero-current level. (C) Summary of the current magnitudes activated in response to RTx relative to saturating 2-APB at +60 mV from experiments as in (B). Values for individual oocytes are shown as open circles and mean ±S.E.M. as squares (n = 3–6). (D) Side view of a cartoon representation of the transmembrane domain of a mTRPV3 subunit (apo, closed structure, PDB: 6DVW) (Singh et al., 2018). The side-chains of residues that enable strong RTx activation when mutated are shown in dark blue, for those that enable weak RTx activation in light blue and for mutations without effect in light grey. Residues within the RTx-binding pocket that were mutated in the 6M construct are highlighted in purple. Helices are shown with 20% transparency, to visualize all side-chains.