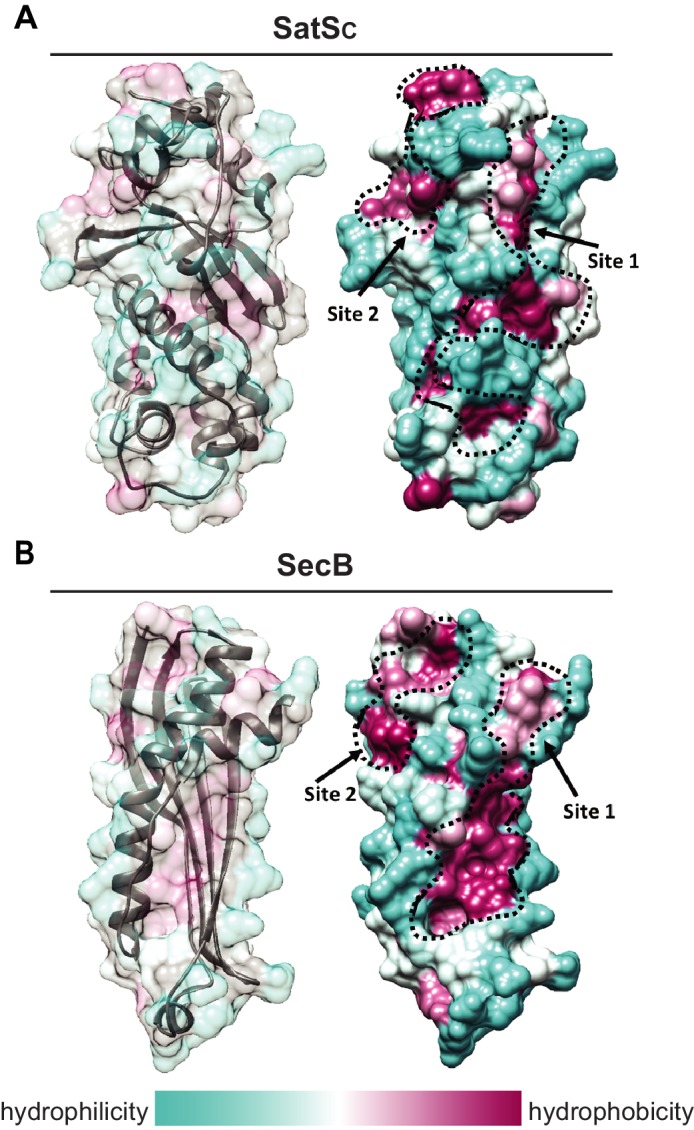
Figure 8. SatS has a new fold and hydrophobic grooves that share similarity with the preprotein binding sites of the SecB chaperone.
(A) The overall secondary structure of SatSC. The hydrophilicity of SatSC is a colored gradient from cyan (hydrophilic) to maroon (hydrophobic). SatSC exposes ~2,900 Å2 of hydrophobic surface. The predicted primary and secondary polypeptide binding site(s) are delineated. (B) The overall secondary structure of SecB monomer (PDB ID:1QYN) (Dekker et al., 2003). The hydrophilicity of SecB is a colored gradient from cyan (hydrophilic) to maroon (hydrophobic). The primary and secondary client binding site(s) are delineated. Each SecB monomer exposes ~1,900 Å2 of hydrophobic surface for client protein interactions (Huang et al., 2016). Molecular graphics and analyses were performed with the UCSF Chimera package (Petersen et al., 2011).