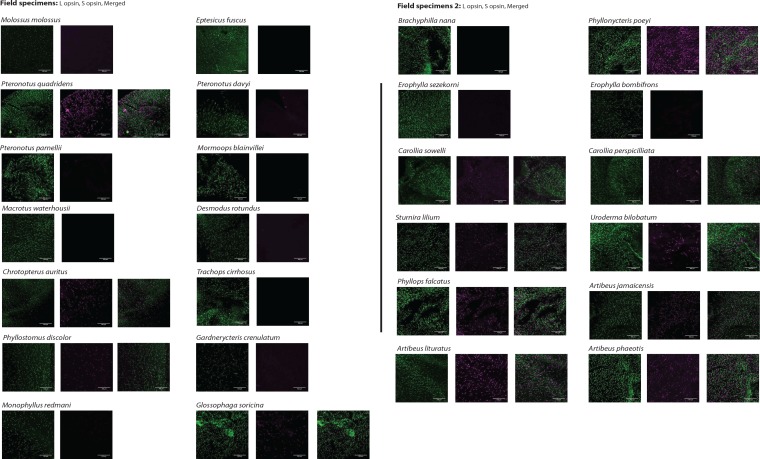
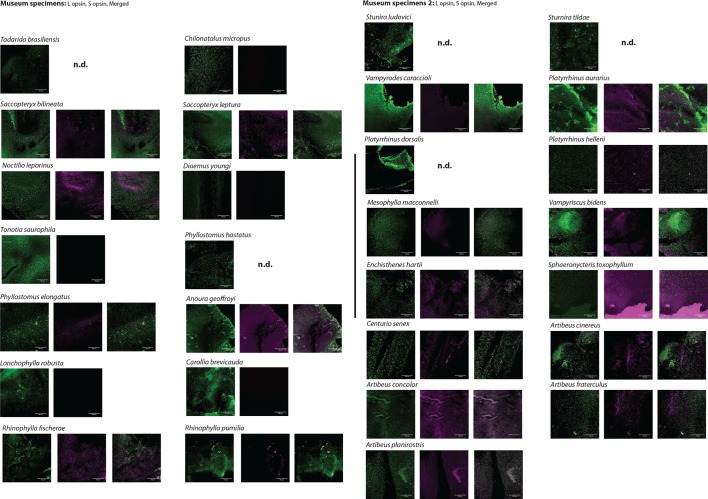
Figure 1. Distribution of an intact open reading frame (ORF), mRNA transcript, and protein for the OPN1SW, OPN1LW, and RHO photopigments in ecologically diverse noctilionoid bats.
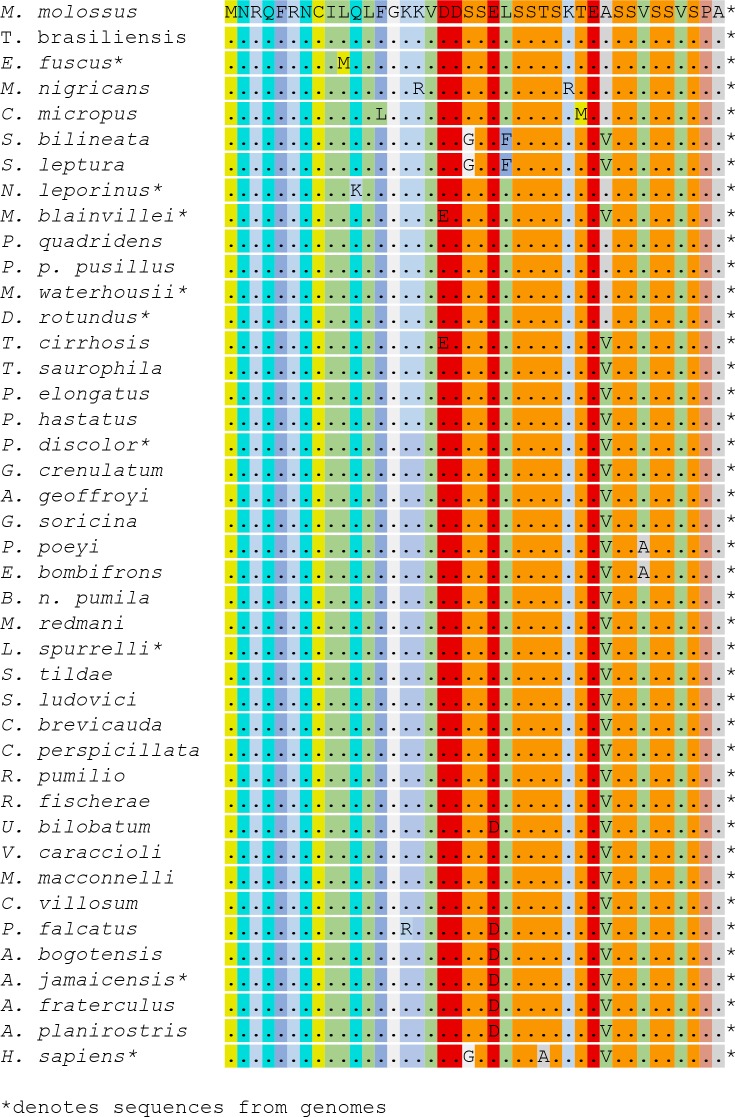
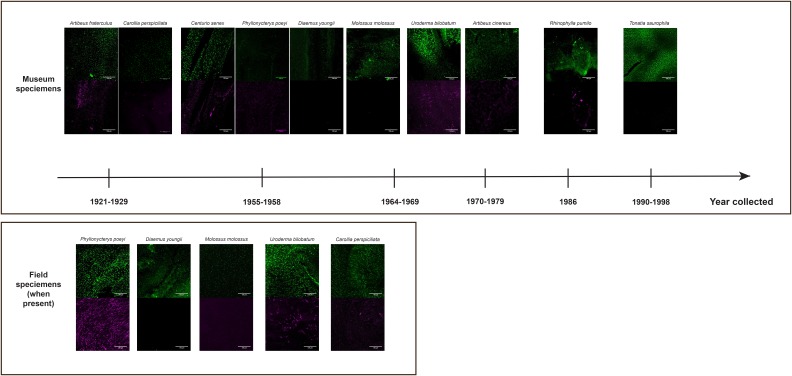
The composition of species diet follows Rojas et al. (2018), dietary types are indicated with the following symbols: invertebrates – moth, vertebrates – frog, fruit – fruit and nectar/pollen – flower. The species phylogeny follows Rojas et al. (2016) and Shi and Rabosky (2015). Vertical black bars, from left to right, indicate: (1) Noctilionoidea, (2) Phyllostomidae, (3) and Stenodermatinae. RNA-Seq data was generated to both infer the presence of an intact ORF (in combination with genomic and PCR sequence data) and to determine the presence of an expressed mRNA transcript. The presence of an intact ORF and mRNA transcript for RHO was verified across all transcriptomes. The presence/absence of a protein product for S- and L-opsins was assayed by IHC on flat mounted retinas. The presence of an intact ORF, mRNA, and protein are indicated by a filled color marker (OPN1SW – purple, OPN1LW – green and RHO – blue), and its absence by a white marker. Missing data (i.e. species for which we were unable to obtain tissue) are indicated with a grey marker with grey outline. Mismatches between intact ORFs and transcripts, or between transcripts and protein data are indicated by an inequality symbol. Note: OPN1SW protein assays for P. quadridens revealed polymorphisms within the sample, and we recorded positive OPN1SW assays in some P. poeyi individuals despite an apparent disrupted ORF. Finally, a grey marker with no outline indicates the failure of protein assay for some species represented by museum specimens (Tadarida brasiliensis, Phyllostomus hastatus, Sturnira tildae, Sturnira ludovici, Platyrrhinus dorsalis and Chiroderma villosum).