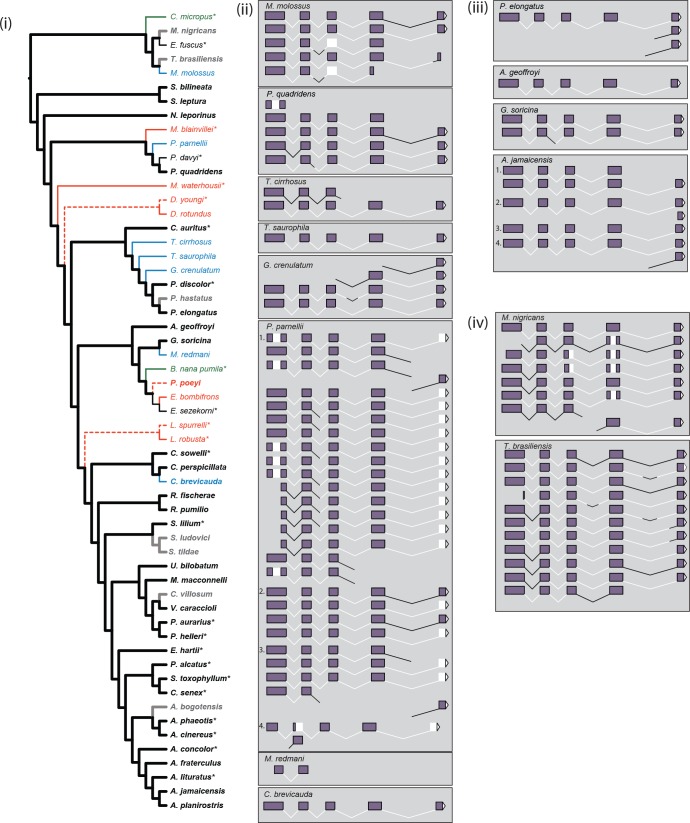
Figure 4. Inferred parallel losses of S-opsins mapped on to the species phylogeny and exonic content of reconstructed mRNA.
(i) Taxa and branches are colored as follows: presence of protein – black; presence of intact ORF but no protein – green; presence of mRNA but absence of protein – blue; evidence of pseudogenization (disrupted ORF) – red; protein status not determined – grey. Weight of branches indicates: inferred presence of protein – heavy; inferred absence of protein – light; protein absence based on evidence of gene loss but not confirmed by IHC – dashed light. We were either not able to recover mRNA, or preserved material was not available, for species marked with ‘*’, evidence for ORF status for Diaemus youngi taken from Kries et al., 2018. The species phylogeny follows Rojas et al., 2016 and Shi and Rabosky, 2015. (ii) Reconstructed mRNA transcript variants of seven species (M. molossus, T. cirrhosus, T. saurophila, G. crenulatum, P. parnellii, M. redmani, and C. brevicauda) with OPN1SW mRNA present but no detected protein, and P. quadridens for which presence of detected protein varied across individuals. The four biological replicates of P. parnellii are numbered 1–4. Sections of the mRNA are indicated as follows: exons 1–5 – purple filled boxes; introns 1–4 – black lines; the 3’UTR – white filled triangle; and missing regions – white regions. (iii) Reconstructed mRNA transcript variants of four species (P. elongatus, A. geoffroyi, G. soricina and A. jamaicensis) with OPN1SW mRNA present and detected protein. The four biological replicates of A. jamaicensis are numbered 1–4. Sections of the mRNA are indicated as above. (iv) Reconstructed mRNA transcript variants of two species (M. nigricans and T. brasiliensis) with OPN1SW mRNA present but protein status not determined. Sections of the mRNA are indicated as above.