Figure 1.
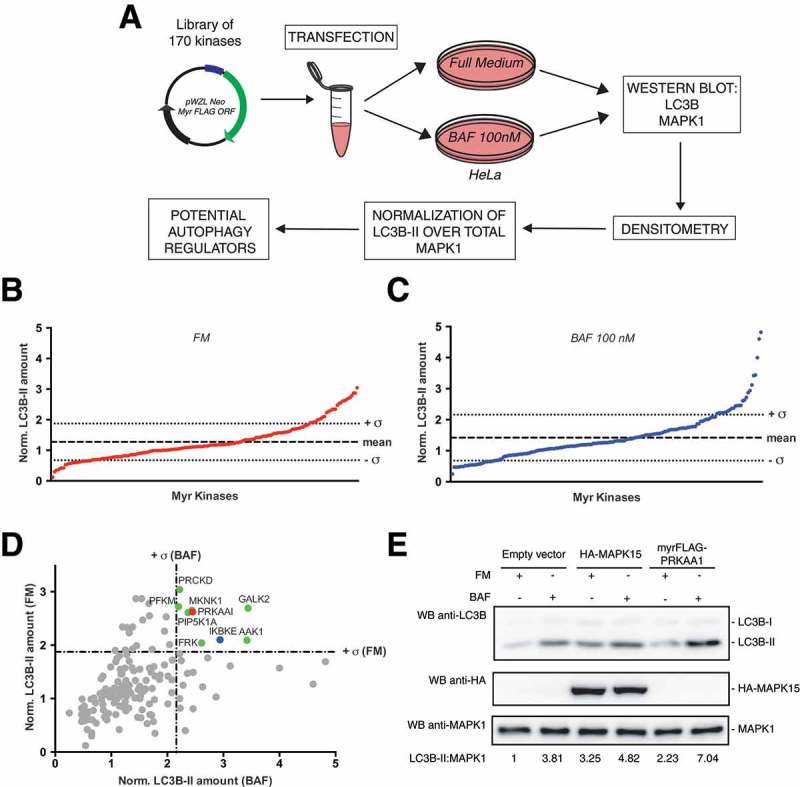
Screening of a library of MYR kinases to identify regulators of the autophagic process. (a) Schematic representation of the experimental approach used to perform the screening of 170 constitutively active kinases for their ability to modulate the autophagic process, in HeLa cells. (b) Normalized LC3B-II levels, obtained by western blot (WB) analysis, of samples transfected with each activated kinase, in Full Medium (FM) conditions. The graph shows also the mean value of the screening and its standard deviation (σ). (c) Same as in (b), but after 1 h of 100 nM bafilomycin A1 (BAF) treatment. (d) Integration of results shown in (b) and (c), to show potential positive regulators of the autophagic flux, identified as kinases inducing normalized LC3B-II values higher than + σ in both FM and BAF conditions (in colors). (e) WB analysis to evaluate autophagic flux of HeLa cells upon myrFLAG-PRKAA1 overexpression. Transfection with an empty vector was used as a negative control. Transfection with HA-MAPK15 was used as a positive control for activation of the autophagic flux. Where indicated, 1 h treatment with 100 nM BAF was performed. Densitometric analysis of LC3B-II levels, normalized by the corresponding MAPK1 levels (used as loading control), is also shown. Results from one experiment, representative of 3 independent experiments (n = 3) are shown.
