Abstract
Purpose:
Transcriptomic profiling can shed light on the biology of SCBC, nominating biomarkers and novel therapeutic targets.
Experimental Design:
Sixty-three SCBC patients had small cell histology confirmed and quantified by a genitourinary pathologist. Gene expression profiling was performed for 39 primary tumor samples, 1 metastatic sample, and 6 adjacent normal urothelium samples (46 total) from the same cohort. Protein levels of differentially expressed therapeutic targets, DLL3 and PDL1, and also CD56 and ASCL1, were confirmed by IHC. A SCBC PDX model was utilized to assess in vivo efficacy of DLL3-targeting antibody-drug conjugate (ADC).
Results:
Unsupervised hierarchical clustering of 46 samples produced 4 clusters that correlated with clinical phenotypes. Patients whose tumors had the most “normal-like” pattern of gene expression had longer OS compared to the other 3 clusters while patients with the most “metastasis-like” pattern had the shortest OS (p=0.047). Expression of DLL3, PDL1, ASCL1 and CD56 was confirmed by IHC in 68%, 30%, 52% and 81% of tissue samples, respectively. In a multivariate analysis, DLL3 protein expression on >10% and CD56 expression on >30% of tumor cells were both prognostic of shorter OS (p=0.03 each). A DLL3-targeting ADC showed durable anti-tumor efficacy in a SCBC PDX model.
Conclusions:
Gene expression patterns in SCBC are associated with distinct clinical phenotypes ranging from more indolent to aggressive disease. Overexpression of DLL3 mRNA and protein is common in SCBC and correlates with shorter OS. A DLL3-targeted ADC demonstrated in vivo efficacy superior to chemotherapy in a PDX model of SCBC.
Introduction
Small cell bladder cancer (SCBC) accounts for approximately 2–5% of all bladder tumors1 and recent data suggest the ‘neuronal’ subtype may be more common.2,3 SCBC is associated with aggressive disease characterized by early progression and metastases. SCBC biology is poorly understood, but its clinical behavior shares similarities with small cell and neuroendocrine tumors of other primary sites, such as small cell lung cancer (SCLC).4,5 It is estimated that 38–70% of SCBC exhibit coexisting non-small cell carcinoma, most commonly invasive and/or in situ urothelial carcinoma (UC).6,7 No standard of care based on randomized clinical trials exists for advanced SCBC, and treatments have been extrapolated from SCLC and bladder UC.8–11 Several retrospective series7,12–15 and small prospective trials11,16 have assessed treatment patterns and efficacy in SCBC. In general, early relapses are common with poor overall outcomes.1,7,19 More effective therapies and more refined prognostic biomarkers are needed in SCBC.
SCBC is underrepresented in the TCGA, leading to separate studies exploring its unique genomic landscape.20,21 New tumor biomarkers and therapeutic targets are emerging through extrapolation from other neuroendocrine malignancies. One example is delta-like protein 3 (DLL3), a Notch pathway ligand overexpressed on the surface of SCLC cells and other neuroendocrine malignancies.22,23 This finding led to the development of a DLL3-targeted antibody-drug conjugate (ADC),24 Rovalpituzumab tesirine (Rova-T), consisting of a DLL3-targeted monoclonal antibody conjugated to a DNA-damaging pyrrolobenzodiazepine (PBD) dimer toxin (Abbvie Stemcentrx, Inc.) with demonstrated efficacy in preclinical23 and in an early phase clinical trials.25 Program death ligand 1 (PDL1), expressed on immune and/or tumor cells is targeted by checkpoint inhibitors, FDA-approved in UC.
Previously reported data in SCLC involving both gene expression profiling26–28 and clinical evaluation of a DLL3-targeting agent led us to consider analogous approaches in SCBC. We hypothesized that gene and protein expression analysis of SCBC samples would validate potential prognostic biomarkers and treatment targets, e.g. DLL3, in SCBC. Our primary objective was to identify histologic biomarkers and novel gene expression classifiers to inform patient selection and clinical trial designs in SCBC. We further sought supporting evidence for the efficacy of novel agents active against targets of interest in pre-clinical models of SCBC.
Methods
Patient Selection and ClinicoPathologic Review
A total of 63 patients with SCBC and available clinical data, seen at Cleveland Clinic from 1993 to 2016, were identified based on pathology records. Clinical and pathologic characteristics, treatment patterns, response and outcomes data were collected for all 63 patients. All tissues were independently reviewed for this analysis by an experienced genitourinary pathologist who confirmed the diagnosis of SCBC and quantitated the small cell component of each tumor (SC%). The study was approved by the Cleveland Clinic Institutional Review Board and was conducted in accordance with guidelines laid out in the Declaration of Helsinki. Written consent from subjects was not required (waiver of consent was granted due to the retrospective nature of the study).
Gene Expression Analysis
Sufficient tissue for gene expression analysis was available for 39 patients utilizing the HTG EdgeSeq Oncology Biomarker Panel Assay (HTG Molecular Diagnostics, Tucson AZ) with probes for 2560 genes validated for in situ expression profiling of FFPE archived specimens.29,30 Other methodologies (RNAseq, microarrays) were excluded due to low input amounts and a requirement for high quality RNA extraction. Gene expression analysis was performed on a total of 46 tissue samples: 39 primary SCBC tumor samples, 1 metastatic sample, and 6 normal bladder tissue samples (from the same 39 patient cohort). The EdgeSeq assay is a targeted NGS approach that produces dispersed probe read counts amenable to RNAseq analysis pipelines.31 Analysis was performed via the RNAseq workflow (Partek Genomics Suite). R (version 3.2.3) was used for statistical analysis of expression data. FDR correction (FDR <5%) and adjusted p values (p<0.05) were used for selection of top differentially expressed genes; candidates were subsequently validated by IHC. All 39 patients included in the gene expression analysis had tissue samples available for immunohistochemistry analyses. Hierarchical clustering was performed to determine Euclidian distance between 46 tissue samples based on the expression of 2560 genes in the EdgeSeq OBP Assay. Kaplan-Meier and log rank proportional hazards testing were used for survival analysis. One-way analysis of variance (ANOVA) was used to detect differentially expressed genes among tumor sample clusters. Paired t-test was used to detect differentially expressed genes between tumor and adjacent normal samples for the six patients with matched samples.
Immunohistochemistry Analyses
Of 63 patients, 53 had tumor tissue samples assessed for DLL3 protein expression via validated Ventana IHC Assay (Ventana Medical Systems, Tucson, AZ) with anti-DLL3 antibody (SC16.65, Abbvie Stemcentrx, South San Francisco, CA).25 DLL3 expression was defined as percentage of tumor cells in a tissue sample that stained positive for DLL3. PDL1 staining was assessed in the 53-patient cohort using both SP263 and SP142 anti-PDL1 antibodies (Ventana, Tucson, AZ). Positive PDL1 expression was defined as ≥1% of tumor infiltrating cells (IC) staining positive for PDL1 using either antibody, based on manufacturer specifications.32,33 The pathologist who interpreted PDL1 staining had received training on both PDL1 assays. Finally, within the 53-patient subset, 52 patients with adequate tissue had IHC staining for neuroendocrine markers ASCL1 and CD56 using Abbvie Stemcentrx SC72.201 and DAKO (123C3) antibodies, respectively. Simultaneous co-staining of biomarkers was not technically feasible. Spearman correlations for associations among biomarkers as well as between biomarkers and relevant patient characteristics, including outcomes, were assessed.
Outcomes Analyses
Univariate and multivariate analyses were used to identify relevant pathologic characteristics and histologic biomarkers among the predictors of overall survival (OS) measured from diagnosis to time of death, progression free survival (PFS) measured from diagnosis to the first recurrence/progression or death, and time to progression (TTP) measured from diagnosis to recurrence/progression with death in the absence of recurrence/progression considered a competing risk. OS, PFS and TTP were also separately calculated for patients who underwent cystectomy; in those, endpoints were measured from the time of cystectomy. Cox proportional hazards models (for OS and PFS) and Fine and Gray model (for TTP) were used in the univariate and multivariate analyses of clinical outcomes. A recursive portioning algorithm was used to identify cutpoints for biomarker expression (DLL3, PDL1, ASCL1, CD56 and SC%) that were associated with differences in outcomes for patients above and below these thresholds.
Patient derived xenograft (PDX) testing
The in vivo efficacy of a DLL3-targeting agent was additionally assessed in a PDX model. BL100 PDX was developed from a cystectomy specimen of a high-grade neuroendocrine bladder carcinoma of a 67-year old Caucasian female after informed consent. BL100 was propagated in 5–7 week old female non-obese diabetic-severe combined immunodeficiency (NOD/SCID) mice (Charles River Laboratories) by subcutaneous implantation of dissociated cells into a single site near the lower mammary fat pad. Animal health was monitored daily, and mouse weights and tumor volumes were measured weekly. All animal studies were approved by the Stemcentrx Institutional Animal Care and Use Committee (IACUC) (Protocols SCAR-3–2008 and SCAR-5–2009) and performed in accordance with American Association for Laboratory Animal Science and AVMA Guidelines. Eight mice were randomized per treatment group so that each treatment group had average tumor volumes of 140–200 mm3. Tumors were measured with digital calipers using two dimensions, long and short axis (mm), and tumor volume (mm3) was calculated as the volume of a prolate ellipsoid: (0.5 x long axis x short axis2). Each group was treated intraperitoneally with either vehicle control, a single dose of cisplatin (5 mg/kg) and etoposide (8 mg/kg) for days 1–3, a single dose of SC16LD6.5 (Rova-T) at 1.6 mg/kg or a single dose of isotype control ADC (HuIgG1LD6.5) at 1.6 mg/kg. The RovaT dose used (1.6mg/kg) was selected from 3 doses of RovaT that were tested on the PDX model (0.4, 0.8 and 1.6 mg/kg). The data for 1.6 mg/kg dose was included because tumors from that cohort were used in the limiting dilution assay to determine impact on tumor initiating cell frequency (described below). The two lower doses were additionally not found to be efficacious. Tumor volumes were assessed weekly.
In vivo limiting dilution assays
To assess whether treatment targets tumor initiating cells (TIC), further experiments were performed. Tumors were removed after euthanasia seven days post-treatment of two representative mice from each treatment group. Cohorts of ten mice per group were injected with decreasing numbers of isolated BL100 tumor cells previously treated with either vehicle, isotype control or Rova-T and ranging from 3,000 to 100 cells per recipient animal. Mice were scored positive for tumor growth if tumors exceeded 200mm3. This serial transplantation of cells from treated mice allowed for the estimation of residual TIC frequency using Poisson distribution statistics. Estimates of TIC frequency were calculated using the L-Calc software package (Stem Cell Technologies) to apply Poisson distribution analysis to the frequencies of tumor-negative mice at each injected cell number.
Results
Clinical, Pathologic, and Treatment Characteristics
In the overall 63-patient cohort, median age was 71, 83% were men, 51% had ECOG Performance Status 0–1, 70% were current/former smokers; 41 patients underwent radical cystectomy and their pathologic characteristics are shown in Table 1. The subset of 39 patients whose tissue samples were available for gene expression analysis had similar baseline characteristics to the overall cohort [Table 1]. Among all 63 patients, SCBC was diagnosed on TURBT for 48 and on cystectomy for 15. At initial diagnosis, 6 patients had distant metastases and 44% (18/41) of patients who had radical cystectomy with lymph node dissection had nodal metastasis. The full description of diagnosis and treatment patterns as well as most commonly used treatment regimens is available in Supplementary materials [Supplementary Figure S1, Supplementary Table 1].
Table 1:
Baseline Patient Characteristics in the Overall Cohort and Among Patients with Gene Expression Data (Left) and Pathologic Characteristics Among Patients with Cystectomies (Right)
| Variable | All Patients (N = 63) |
Patients with Data on Gene Expression (N = 39) |
Pathologic Features of Cystectomy Patients |
All Cystectomy Patients (N = 41) |
Cystectomy Patients with Data on Gene Expression (N=27) |
|---|---|---|---|---|---|
| Median Age | 71 (39–90) | 70 (46–89) | Tumor
Location Bladder Bladder and Ureter Bladder and Urethra No tumor (T0) |
37 (90%) 2 (5%) 1 (2%) 1 (2%) |
25 (92%) 1 (4%) 0 1 (4%) |
| ECOG PS at Diagnosis 0: 1: 2: 3: Unknown |
22 (35%) 10 (16%) 4 (6%) 3 (5%) 24 (38%) |
11 (28%) 8 (21%) 3 (8%) 3 (8%) 14 (36%) |
Pathologic T-stage T0-T2 T3-T4 |
15 (37%) 26 (63%) |
9 (33%) 18 (67%) |
| Gender Male Female |
52 (83%) 11 (17%) |
33 (85%) 6 (15%) |
Pathologic
N-stage N0: N1-N3: |
23 (56%) 18 (44%) |
17 (63%) 10 (37%) |
| Current/Former Smoker Yes No Unknown |
44 (70%) 15 (24%) 4 (6%) |
29 (74%) 6 (16%) 4 (10%) |
Surgical
Margins Positive Negative Unknown |
8 (20%) 29 (70%) 4 (10%) |
6 (22%) 18 (67%) 3 (11%) |
| Hydronephrosis Yes No Unknown |
9 (14%) 53 (84%) 1 (2%) |
7 (18%) 32 (82%) 0 |
Carcinoma in Situ
(CIS) Present Absent Unknown |
24 (59%) 16 (39%) 1 (2%) |
14 (52%) 12 (44%) 1 (4%) |
| Lymphovascular Invasion
(LVI) Yes No Unknown |
17 (41%) 10 (24%) 14 (35%) |
13 (48%) 5 (19%) 9 (33%) |
Gene Expression Profiling
Gene expression profiling was performed on 39 primary SCBC tumor samples with sufficient tissue. In six of those patients, gene expression profiling was also performed on matched adjacent “normal-appearing” urothelial tissue. Analysis of a single metastatic tissue was also performed [Figure 1A]. Principle components analysis of gene expression parameters for each of the 46 analyzed samples is depicted in Figure 1B and displays grouping of expression profiles by tissue origin (tumor, normal, and metastatic).
Figure 1:
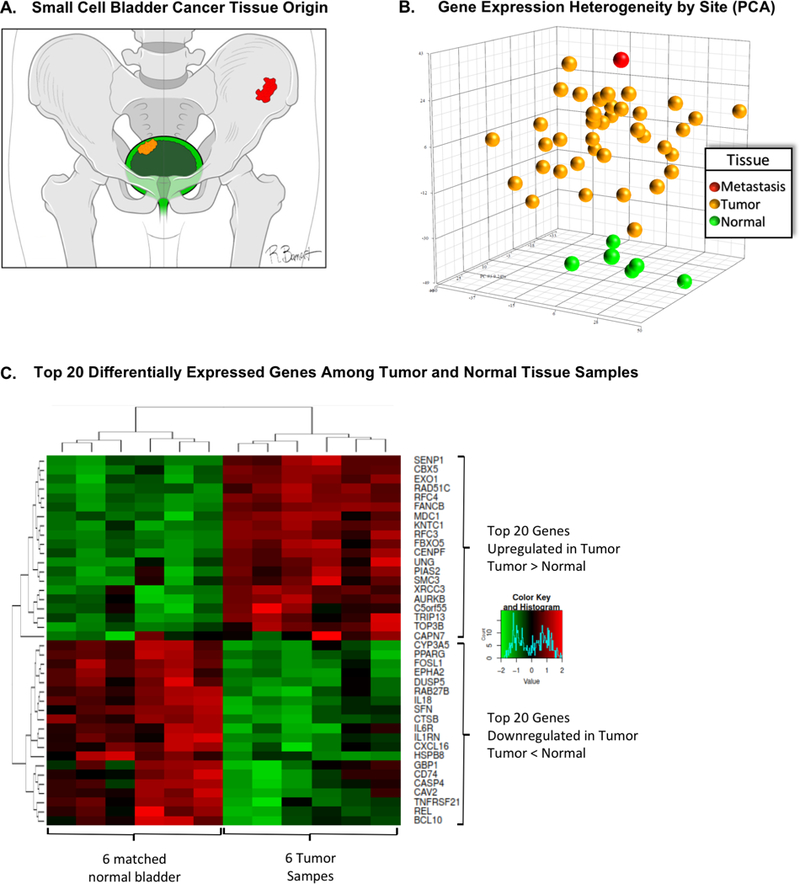
A: Diagram of the tissues of origin utilized in the gene expression analysis. Green represents normal urothelial tissue samples, yellow represents tissue samples from primary bladder tumor, and red represents tissue from metastatic sample (osseous metastasis). B: Three-dimensional representation of the principal component analysis which represents tissue gene expression heterogeneity across 2560 genes as a Euclidian distance in three principal components. This displays a similarity in gene expression among normal urothelial samples (green), which stand apart from the gene expression similarity observed across primary small cell bladder tumors (yellow), and finally apart from gene expression in the one metastatic sample (red) which is one of the gene expression outliers. C: Differential gene expression analysis comparing gene expression in 6 matched tumor and normal urothelial samples from the same patients. Top of the panel displays the top 20 genes with increased expression in tumor relative to normal samples, while the bottom panel displays the top 20 genes with greater expression in normal samples as compared to tumor samples
Unsupervised hierarchical clustering of gene expression patterns from all 46 samples produced 4 distinct gene expression clusters [Figure 2A]. Consensus (K-means) clustering confirmed 4 distinct subsets [Supplementary Figure S2]. Importantly, membership within clusters was associated with discrete clinical phenotypes that correlated with OS [Figure 2C]. Patients with tumor samples in cluster 2, containing all 6 normal urothelial tissue samples, had the most favorable clinical phenotype. The co-clustered tumor and normal samples originated from different patients, suggesting greater similarity between clinically indolent tumors and normal tissues than between tumor and normal tissues from a single individual. Patients in cluster 2 did not develop metastasis. On the other hand, patients in Cluster 3 whose tumors had a gene expression pattern that grouped them with the metastatic tumor sample, were more likely to have metastasis and had shorter median OS (6 mos) compared to patients in the other three clusters (Kaplan-Meier log-rank p=0.047). The top 20 differentially expressed genes among the 4 clusters (associated with the highest ANOVA p-values) are shown in Figure 2B. Gene set enrichment analysis (GSEA) for top 200 differentially expressed genes among the 4 clusters demonstrated distinct dominant biological networks (Supplementary Figure S3).
Figure 2:
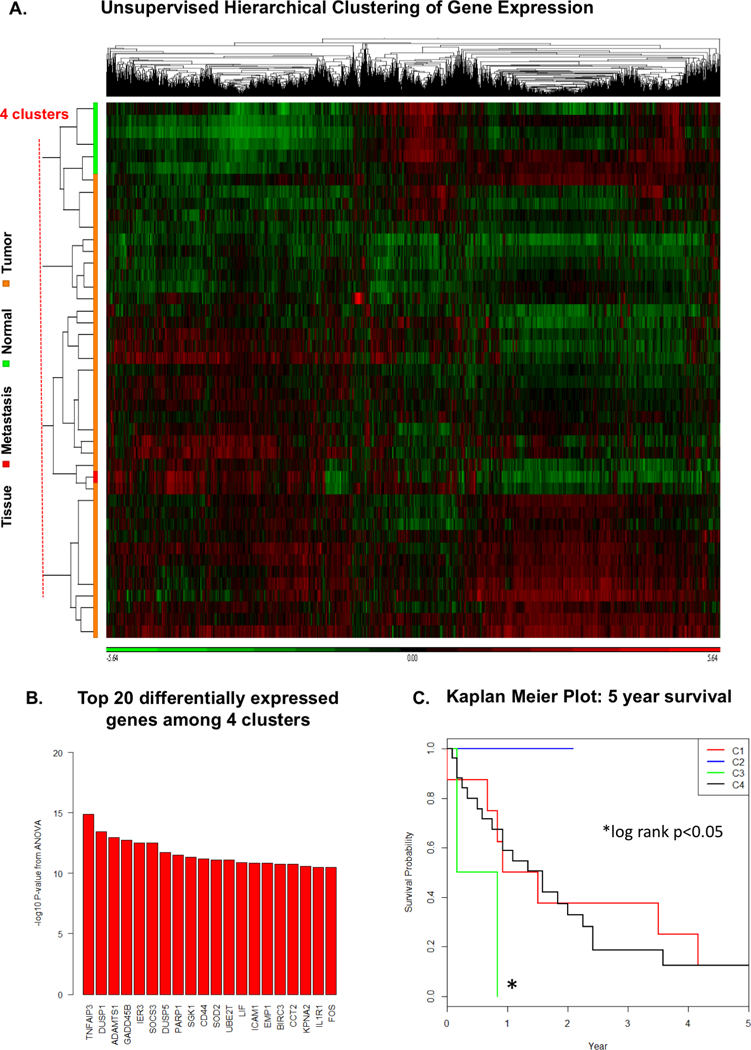
A: Unsupervised hierarchical clustering separates individual tissue samples (rows on vertical axis), based on relative overexpression (green) or underexpression (red) of individual genes on the horizontal axis, into 4 distinct clusters of similar gene expression patterns. B: Top differentially expressed genes among 4 clusters C: Kaplan-Meier plot of overall survival (OS) for patients across 4 identified gene expression clusters. Gene expression clusters correlate with distinct clinical phenotypes as patients with tumors in most “normal-like” C2 had superior OS whereas patients in the most “metastasis-like” C3 have shorter OS (median OS 6.0 months) compared to the other 3 clusters (log rank p = 0.047)
Among the six matched pairs of tumor and adjacent normal tissue from the same patient, the differential gene expression analysis using matched pairs resulted in a total of 583 differentially expressed genes with FDR < 0.05. The top differentially expressed genes (based on p-value) for the tumor vs. normal tissue comparison and the associated heatmap are shown in Figure 1C, while the GSEA plots are available in Supplementary Figure S4. EZH2, which is overexpressed or mutated in numerous malignancies and targeted by an agent being tested in clinical trials34 was significantly overexpressed in the six SCBC tumors compared to normal tissue samples (p = 0.0003) [Figure 3D]. This finding is consistent with prior reports of EZH2 and associated polycomb repressor group (PRC) dysregulation in SCLC.35,36
Figure 3:
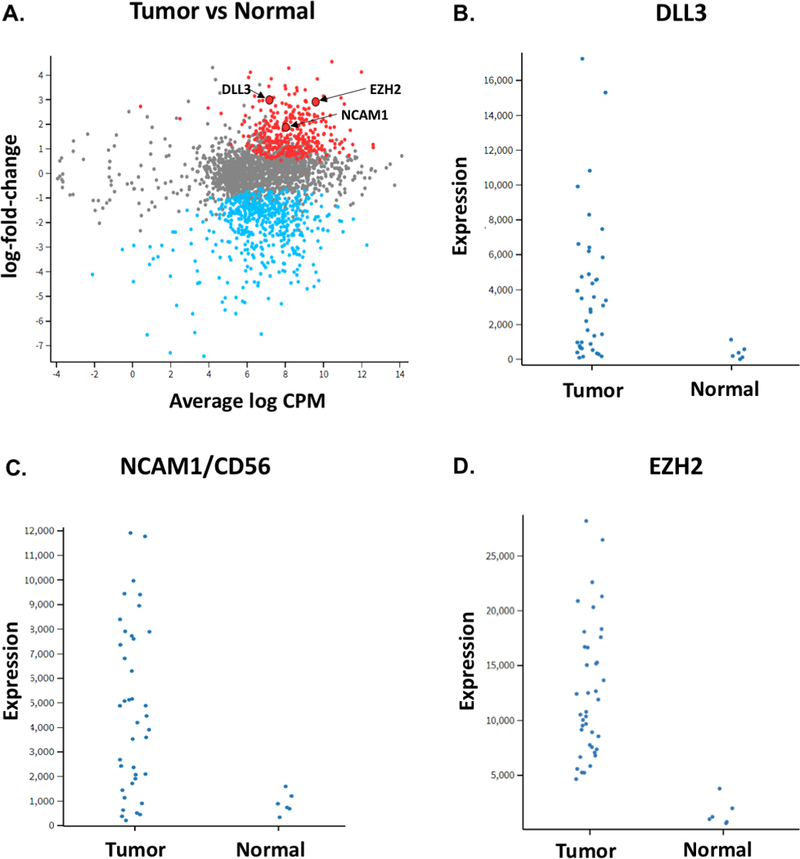
(A) Gene expression log fold change vs average log of counts per million (CPM) across samples; significantly differentially expressed genes (log fold change >1.0, adj p<0.05) are displayed in red if upregulated and blue if downregulated in tumors vs normal. Arrows highlight genes of interest. Plots for (B) DLL3, (C) NCAM1/CD56 and (D) EZH2 display gene expression in individual tumor and available normal samples
Among genes with significantly higher mRNA expression in SCBC tumor samples relative to normal urothelial tissue, DLL3 (p=0.02) and NCAM1(CD56) (p=0.007) represented potential biomarkers of interest given their prior association with other small cell or neuroendocrine malignancies; and in the case of DLL3, the availability of Rova-T [Figure 3A-C]. Expression of DLL3, CD56 and other proteins was then assessed in patients with available tumor samples (see below). Among all 2560 genes in the HTG EdgeSeq panel, DLL3 protein expression was most strongly correlated with expression of DLL3 mRNA (Supplementary Figure S5), suggesting that regulation of DLL3 expression occurs primarily at the transcriptional rather than translational level.
Biomarker Expression and Small Cell Component
Gene expression analysis identified candidate proteins for further investigation as relevant biomarkers of interest. All 63 SCBC tumors had confirmed small cell component (SC%) with a range of 5%−100%; 59% of tumors had SC% of 100% (pure small cell histology) and 79% had SC% of ≥50% (small cell as dominant histology). Among patients with a mixed histology, UC was the most common non-small cell histology. Among 39 patients with available gene expression data, SC% had a significant correlation with mRNA expression of DLL3 (r=0.68, p=0.02) and CD56 (r=0.42, p=0.008).
In 53 patients with available tissue specimens for IHC of DLL3 (therapeutic target of Rova-T) and PDL1 (therapeutic target of checkpoint inhibitors), DLL3 protein expression (≥1% of tumor cells) on IHC was noted in 68% of patients, with 58% having expression in >10% of tumor cells [Figure 4]. The median percentage of DLL3-positive tumor cells among all samples was 40% (range 0–100%). There was a positive correlation between percentage of DLL3-positive tumor cells and SC% of tumor (Spearman r=0.33, p=0.01), DLL3 mRNA expression (r=0.70, p=1.0×10−7) and with mRNA expression of neuroendocrine markers and Notch pathway proteins including DLL4 (r=0.44, p=0.004) and CHGA (r=0.55, p=0.0003). A negative correlation was noted between DLL3 IHC and mRNA expression of Notch1 (r=−0.47, p=0.002), RB1 (r=−0.48, p=0.002) and EZH2 (r=−0.29, p=0.07). PDL1 protein expression via IHC was only noted on tumor-infiltrating immune cells and not on tumor cells. In 30% of available samples, PDL1 expression was seen on ≥1% of tumor IC using either SP263 or SP142 anti-PDL1 antibodies (21% for SP263 alone, 2% for SP142 alone and 7% for both SP263 and SP142). However, no case reached previously defined criteria for either high or positive expression for SP263 or SP142 antibody, respectively due to lack of tumor cell or IC staining in the remaining tumor microenvironment. No significant correlation was noted between PDL1 protein expression and either DLL3 protein expression, SC%, or outcomes.
Figure 4:
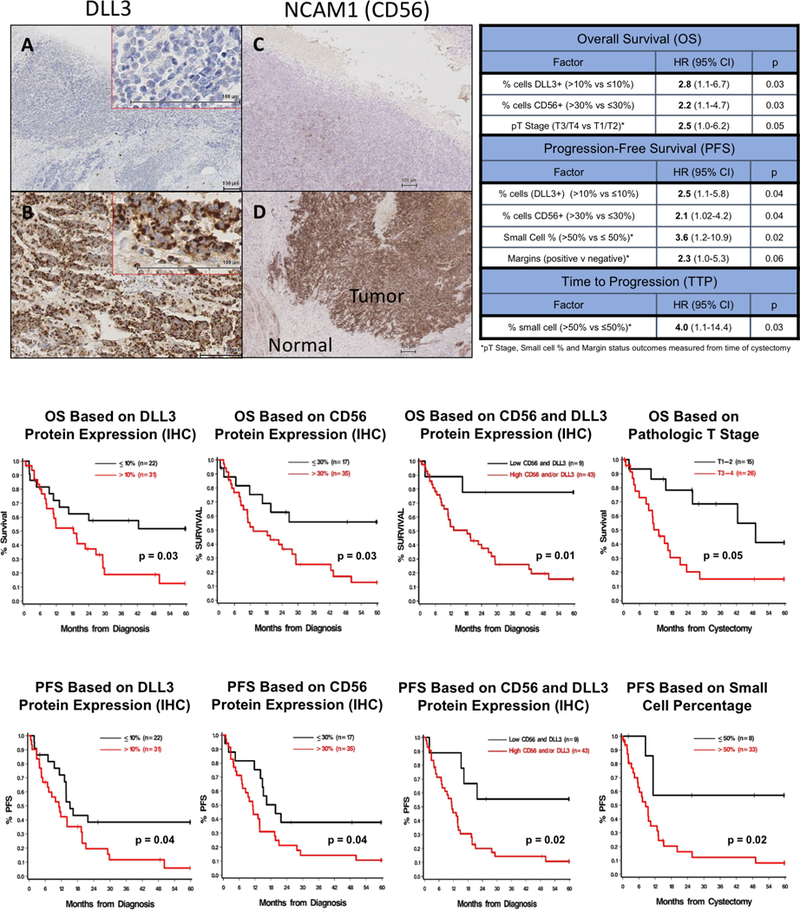
Top Left: Immunohistochemistry for DLL3 Expression: (A) Negative control and (B) tissue with 95% of tumor cells expressing DLL3. IHC for CD56 Expression: (C) Negative control and (D) tissue with 70% of tumor cells expressing CD56 Top Right: Multivariate analyses of tumor characteristics associated with relevant clinical outcomes including overall survival, progression-free survival and time to progression. Bottom: Kaplan-Meier plots and associated p-values for overall survival and progression-free survival based on pre-determined cutpoints for tumor markers and other characteristics
Among 53 patients with IHC of DLL3 and PDL1, 52 patients also had tissue samples available for IHC of CD56/NCAM1 and ASCL1(one patient excluded due to artifact). Positive cell membrane expression of CD56/NCAM1 protein (≥1% of tumor cells) was observed in 81% of tumors [Figure 4]. The median percentage of CD56-positive cells in tumor tissue was 65% (range 0–100%), and lower CD56 expression was noted in patients older than age 70 (p=0.05). CD56/NCAM1 protein expression was correlated with SC% (r=0.26, p=0.06), DLL3 protein expression (r=0.32, p=0.02), and with mRNA expression of neuroendocrine markers including NCAM1 (r=0.61, p=0.00003), DLL4 (r=0.38, p=0.017), Notch1 (r=−0.34, p=0.034) and SYP (r=0.34, p=0.033). Positive expression of ASCL1 transcription factor (≥1% of tumor cells) was observed in 52% of tumor tissues but the median percentage of ASCL1-positive cells within a tumor was only 10% (range 0–100%). ASCL1 protein expression had strong correlation with protein expression of DLL3 (r=0.73, p=0.00001) and moderate correlation with expression of CD56 (r=0.35, p=0.01), and SC% (r=0.34, p=0.01). Significant correlation of ASCL1 protein expression with mRNA expression of several Notch pathway proteins and neuroendocrine markers was observed including: DLL3 (r=0.46, p=0.0035), DLL4 (r=0.60, p=0.00005), CHGA (r=0.41, p=0.01), Notch1 (r=−0.34, p=0.002) and EZH2 (r=−0.31, p=0.05).
Clinical Outcomes and Prognostic Factors
In the overall 63-patient cohort, the median follow-up was 16.6 months from the time of tissue-confirmed diagnosis of SCBC. Median OS was 22.8 months (95%CI 11.9–42.4) and median PFS was 13.7 months (95%CI 11.2–19.4). Multivariate analyses revealed several independent prognostic factors for OS, PFS and TTP in this cohort [Figure 4]. Increased DLL3 protein expression, with a determined cutoff of >10% of cells based on a recursive portioning algorithm, was associated with shorter OS and PFS from diagnosis and shorter OS from cystectomy (p ≤ 0.05 for all). Increased CD56/NCAM1 protein expression (>30% of tumor cells) was similarly associated with shorter PFS (HR: 2.07; 95%CI 1.02–4.20, p=0.04) and OS (HR: 2.23; 95%CI 1.06–4.70, p=0.03). A subset of nine patients whose tumor samples had low expression of both DLL3 (≤10%) and CD56 (≤30%) had significantly longer OS (103.4 vs 18.4 months, p=0.01) and PFS (92.2 vs 11.4 months, p=0.02) relative to patients with high protein expression of either biomarker. On the other hand, increased SC component (>50%) was not associated with OS, but was associated with shorter PFS from cystectomy (p=0.02) and shorter TTP from cystectomy (p=0.03). Neither PDL1 nor ASCL1 protein expression assessed via IHC had association with survival outcomes in this cohort. Pathologic T3/4 stage at cystectomy was associated with shorter OS from the time of cystectomy compared to pT1/T2 (p=0.05). Positive margin status at cystectomy had trend towards shorter PFS from cystectomy, although this finding did not reach statistical significance (p=0.06). Kaplan-Meier plots of OS and PFS for significant prognostic factors are depicted in Figure 4.
Pre-Clinical Efficacy of DLL3-Targeting Agent
With DLL3 protein expression shown to be common in SCBC samples and potentially prognostic of poor outcomes, we pursued assessment of the pre-clinical efficacy in a PDX model of a novel agent (Rova-T) that exploits the expression of DLL3 on the surface of SCBC tumor cells to deliver a cytotoxic agent. FFPE samples of a SCBC PDX model, BL100, were stained by IHC with antibodies specific to DLL3, and shown to express DLL3 on the surface of all tumor cells [Figure 5A]. Rova-T was assessed for in vivo efficacy in these models. As described, following implantation of BL100 tumor cells into four groups of NOD/SCID mice, each group was treated with either vehicle control, a single dose of cisplatin plus etoposide on days 1–3, a single dose of SC16LD6.5 (Rova-T) or an isotype antibody control ADC (HuIgG1LD6.5) [Figure 5B]. Neither vehicle nor cisplatin/etoposide impeded tumor growth, while the isotype ADC showed non-specific response that repressed tumor growth for only ~50 days. A single dose of Rova-T, on the other hand, prevented tumor growth for >100 days, providing durable and specific anti-tumor response [Figure 5C]. No significant treatment-related toxicities were observed in the mice.
Figure 5:
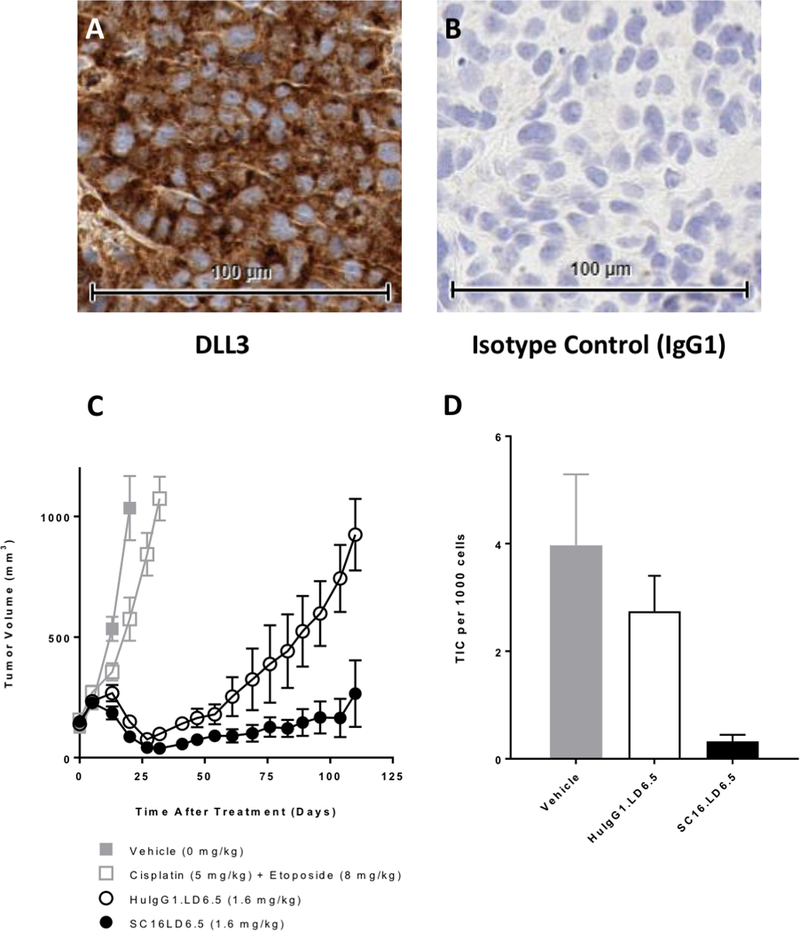
(A) DLL3 immunohistochemistry of FFPE sample of a SCBC patient-derived xenograft (PDX) model and (B) isotype control (IgG1) of same FFPE sample showing lack of staining. (C) Tumor volumes across time in 4 groups of NOD/SCID mice implanted with BL100 tumor cells (SCBC PDX model) and treated with either vehicle, Cisplatin/Etoposide, SC16LD6.5 (antibody-drug conjugate Rova-T), or huIgG1LD6.5 (isotype antibody control ADC) (D) Estimated residual tumor initiating cell (TIC) frequency in tumors previously treated with vehicle, huIgG1LD6.5 or SC16LD6.5, suggesting that TIC suppression by SC16LD6.5 (Rova-T) is the mechanism that induces durable responses to Rova-T treatment
To determine whether Rova-T prevents recurrence by targeting TIC, tumors were harvested from two mice in each treatment group and limiting dilutions of isolated BL100 cells were re-transplanted into cohorts of 10 mice per treatment. Vehicle-treated BL100 PDX tumors had TIC frequency around 1:252 cells, which was reduced to ~1:365 by HuIgG1LD6.5 treatment, and significantly reduced to ~1:3036 by a single dose of Rova-T [Figure 5D]. Collectively, these experiments support durable in vivo efficacy of Rova-T, which effectively targets DLL3-expressing TIC in a SCBC PDX model.
Discussion
We report one of the largest studies of gene expression profiling in SCBC and the first to assess the prognostic value of differentially expressed proteins associated with neuroendocrine differentiation, including DLL3, ASCL1, CD56/NCAM1, and PDL1. We identified a gene expression cluster associated with aggressive biology in SCBC with distinct molecular taxonomy associated with poor prognosis. Additionally, DLL3 and CD56/NCAM1 were identified as negative prognostic biomarkers in SCBC, and DLL3 was further validated as a potential therapeutic target with pre-clinical evidence supporting in vivo efficacy of a DLL3-targeting ADC in a SCBC PDX model. These data have implications for future therapeutic strategies in SCBC.
All patients in this study had small cell histology independently confirmed and quantitatively defined by an experienced genitourinary pathologist. Although SCBC was not included in the original TCGA analysis of bladder cancer published in 2014,37 several recent studies including the updated TCGA analysis published in 2017,2 have added to our understanding of the SCBC genomic landscape. These studies highlight similarities of SCBC to SCLC as well as important differences.20,28,38 Recently published data suggests that SCBC and SCLC have a convergent but distinct pathogenesis, with SCBC having high somatic mutational burden driven by APOBEC mutations--also commonly found in UC--which may precede TP53 and RB1 mutations that typify small cell malignancies.38 The notable high incidence and role of APOBEC mutagenesis is currently unknown and merits further investigation with respect to its relationship with DLL3 expression in SCBC.
Available genomic datasets in bladder cancer have largely focused on the more common urothelial histology. The updated TCGA analysis of 412 muscle-invasive bladder cancers included only 4 tumors with neuroendocrine histology. Three of these 4 tumors clustered with the “neuronal” molecular subtype that included an additional 17 tumors (20 total) without histopathologic features of neuroendocrine origin. The “neuronal” molecular subtype was characterized by typical neuroendocrine markers as well as frequent loss or mutation of TP53 and RB1 and high expression of neuronal differentiation and development genes. Consistent with the known aggressive clinical phenotype of small cell and neuroendocrine bladder cancers, the neuronal subtype had the poorest survival among the five molecular subtypes identified in the updated TCGA analysis.2 These findings support the notion that integrative molecular profiling of tumor samples may complement the histopathologic diagnosis of SCBC, as most neuronal subtype tumors lacked small cell or neuroendocrine histology. Our results further substantiate this premise by demonstrating an association between gene expression signatures and clinical behavior among histologically confirmed SCBC samples. Acknowledging the relatively small number of patients and inherent limitations of retrospective analysis, these findings should be regarded as hypothesis-generating. Comparisons of gene expression data from our analysis with the TCGA dataset and data from additional SCBC cohorts is currently being pursued.
In our analysis, unsupervised hierarchical clustering of gene expression profiles for 39 tumor samples, 6 matched “normal” bladder tissue samples and 1 metastatic site sample revealed four stable gene expression clusters that correlated with clinical phenotypes. We included normal samples in the clustering analysis to assess their inherent similarity and determine the intra-individual transcriptomic differences between tumor and adjacent “normal-appearing” tissues. Recognizing the limitations of assessing the “normal” adjacent tissue in the context of field cancerization and tumor heterogeneity, a number of provocative patterns emerged from this gene expression analysis. For instance, the single metastatic sample clustered closely with the primary tumor sample from the same patient. In contrast, the 6 primary tumors and their 6 matched normal tissue samples did not reliably cluster together. One interpretation is that gene expression among SCBC samples from different individuals may converge on a common transcriptional program, appearing more closely related than expression patterns in matched tumor and normal tissue samples from the same patient. This observation supports prior findings in UC, demonstrating shared early “truncal” events in the course of tumorigenesis and subsequent convergent clonal evolution.39 The similarity of gene expression patterns in the metastatic sample with the primary tumor from which it arose and with another primary tumor from a patient with similarly poor outcome further supports convergent evolution towards more aggressive disease phenotype. On the other hand, tumor samples that clustered closely with normal samples--or had a more “normal-like” pattern of gene expression--demonstrated more indolent clinical course. Taken together, these data suggest that membership in specific gene expression clusters may reflect underlying biology explaining their prognostic value for individual patients. These findings, while provocative, remain to be validated prospectively and are limited in the present dataset by the availability of a single metastatic sample. Identifying the specific genetic determinants of a “metastasis-like” or “normal-like” expression signature in SCBC represents the next logical step in developing risk stratification models to guide management. The differential gene expression appears to be driven by overlapping gene networks, and functional studies are ongoing to better define pathways involved and their role in modulating clinical phenotypes.
We confirmed a strong correlation between mRNA and protein expression of DLL3 and similar findings with CD56/NCAM1. These findings confirmed our hypothesis that regulation of these proteins in SCBC likely occurs at the transcriptional rather than translational level. Our results indicate that the expression of ASCL1 and CD56/NCAM1 proteins in SCBC tumors is also correlated with mRNA expression of several Notch pathway proteins and proteins related to neuroendocrine differentiation such as DLL3, DLL4, Notch 1, CHGA and SYP. Further efforts to better characterize these pathways and the underlying biological mechanisms in SCBC are ongoing.
The expression of DLL3, a promising treatment target in early phase clinical trials in SCLC and other neuroendocrine malignancies, has not been previously reported in SCBC. The majority (68%) of SCBC tumors in our cohort had positive DLL3 protein expression which is similar to SCLC and other solid malignancies, such as melanoma, glioblastoma, and medullary thyroid cancer.40 This is the first study to report increased DLL3 and CD56 expression as independent negative prognostic biomarkers in SCBC associated with shorter PFS and OS in multivariate analyses. Conversely, low simultaneous IHC expression of both DLL3 and CD56 may identify long-term SCBC survivors, as nine patients that met these criteria had median OS exceeding 100 months.
In addition to being a prognostic biomarker in SCBC, DLL3 expression may be an important predictive biomarker and treatment target. A phase I trial of Rova-T, the antibody-drug conjugate that exploits DLL3, in a heavily pre-treated population of patients with SCLC and large-cell neuroendocrine carcinoma showed robust response in patients with limited treatment options.25 A clinical trial of Rova-T in DLL3-expressing solid tumors is ongoing (NCT02709889) and includes SCBC patients in one of the neuroendocrine carcinoma cohorts.41 The high proportion of SCBC tissue samples expressing DLL3 in a high percentage of tumor cells suggest the potential clinical utility of DLL3-targeted agents in this population. Our pre-clinical data demonstrating Rova-T efficacy in DLL3-expressing SCBC PDX models is further supportive. Prior studies have shown Rova-T to have limited efficacy in small cell lung cancer PDX models that did not express DLL3 and no cross-reactivity against DLL4. This suggests that tumor cell killing by Rova-T is mediated through DLL3.23 Furthermore in our analysis, DLL3 expression was found to localize exclusively to the membrane and cytoplasm of the SC tumor component and was not expressed in the urothelial component, endothelium, or in adjacent normal tissue. We therefore hypothesize that DLL3-targeting agents would preferentially target the SC tumor component. If this is confirmed in future studies, therapy selection in SCBC may be driven by tumor SC%.
In the study described here, SCBC PDX models treated with a single dose of Rova-T showed durable repression of tumor growth relative to models treated with cisplatin/etoposide and models treated with humanized IgG control isotype (HuIgGLD6.5) with the same drug-conjugate arm. This partial activity of HuIgGLD6.5, which has also been observed in other PDX models, could be explained by its ability to bind Fc-receptors expressed on myeloid cells. Presence of these myeloid cells in the PDX tumor environment could account for the observed non-specific killing of tumor cells following release of the potent drug warhead in the tumor microenvironment.42 Furthermore, serial dilution experiments suggested that more durable repression of tumor growth with Rova-T relative to the control isotype may be due to more effective targeting of TIC by Rova-T. The data suggest the hypothesis that TIC’s may represent a potential DLL3-driven pathologic mechanism of tumor progression that if adequately targeted with DLL3-specific agents may result in durable responses. Our pre-clinical data and the previously described efficacy of Rova-T in other small cell and neuroendocrine malignancies support clinical trials in SCBC.
Our analyses had a number of limitations. As in all retrospective analyses, the full spectrum of confounding and selection biases are difficult to account for. The heterogeneity of the population that included localized and advanced disease, and diverse diagnostic, follow up and treatment patterns, made meaningful comparisons of subpopulations impractical. The 23-year timespan of the cohort is challenging due to the dynamic diagnostic and treatment paradigms during this period. To some extent, this was offset by the fact that all tissues were independently reviewed and quantified by an experienced specialized pathologist using contemporary standards for SCBC diagnosis. Although co-staining of biomarkers was not technically feasible, IHC of each biomarker was read by an experienced pathologist to reduce inter-observer variability. Microdissection of SC component was not performed prior to EdgeSeq analysis raising the question of SC tissue purity; however, gene expression patterns were analyzed along with SC%, and many of the clinical cases have mixed small cell and urothelial histology. Finally, gene expression analysis was available in 39 of 63 patients raising the possibility of selection bias. However, the comparison of the 39-patient subset to the 63-patient overall cohort did not reveal substantial differences in clinico-pathologic characteristics.
In summary, this was the first analysis to suggest that distinct gene expression patterns in SCBC identify more aggressive or indolent behavior and are associated with outcomes. We also report of the frequency and prognostic value of DLL3, PDL1, CD56 and ASCL1 protein expression in SCBC. This work implicates increased DLL3 expression and increased CD56 expression as negative prognostic biomarkers. DLL3 may additionally be an important therapeutic target given its overexpression specifically on the SC component. We demonstrate the efficacy of a DLL3-targeting agent in a PDX model of SCBC. PDL1 assessment demonstrated lack of tumor cell staining consistent with literature in other neuroendocrine tumors, but relatively low IC expression with potential treatment implications.43 Our findings merit prospective validation and comprise an important step in understanding the biology of SCBC that may inform novel prognostic models, treatment paradigms and clinical trial design.
Supplementary Material
Translational Relevance.
The biology of SCBC is not well defined and effective treatment options are limited. In this study, gene expression clustering separated SCBC tumors into groups associated with more aggressive or more indolent disease. We identified increased gene expression of DLL3 (Delta Like Canonical Notch Ligand 3) in SCBC, and verified increased DLL3 protein expression by immunohistochemistry in the tumor samples. Increased protein expression of DLL3 was associated with shorter overall survival in SCBC. DLL3 was separately assessed as a potential therapeutic target with pre-clinical data demonstrating efficacy of a DLL3-targeting antibody-drug conjugate (ADC) in a SCBC PDX model. Data support the potential utility of transcriptomic analysis in identifying clinically distinct molecular taxonomies in SCBC and nominate DLL3 as a potential therapeutic target. The ability to separate SCBC patients into distinct prognostic groups may inform treatment and clinical trial design in SCBC.
Acknowledgements
The authors would like to thank Sam Williams, Marybeth Pysz, Hanna Ramoth, Tabita Popovici and Andrew Hsieh for their contributions.
This study was supported by the Bioinformatics, Imaging, and Integrated Genomics Shared Resources of the Case Comprehensive Cancer Center (P30 CA043703). This work was supported by a Velosano Foundation Cancer Research Award to OYM.
Footnotes
Conflict of Interest: The remaining authors declare no potential conflicts of interest.
Disclosures: LR Saunders, K Isse and E Bishop are employed by Abbvie Stemcentrx. A Dowlati reports being on the advisory board for Abbvie. P Grivas reports consulting with Merck & Co., BMS, Clovis Oncology, EMD Serono, AstraZeneca, Seattle Genetics, Foundation Medicine, Pfizer, QED Therapeutics, Driver Inc. within the past year which was unrelated to this study.
References
- 1.Fahed E, Hansel DE, Raghavan D, Quinn DI & Dorff TB Small cell bladder cancer: biology and management. Semin. Oncol. 39, 615–618 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Robertson AG et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 171, 540–556.e25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meeks JJ & Lerner SP Molecular Landscape of Non-Muscle Invasive Bladder Cancer. Cancer Cell 32, 550–551 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Cheng L et al. Small cell carcinoma of the urinary bladder: a clinicopathologic analysis of 64 patients. Cancer 101, 957–962 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Mukesh M, Cook N, Hollingdale AE, Ainsworth NL & Russell SG Small cell carcinoma of the urinary bladder: a 15-year retrospective review of treatment and survival in the Anglian Cancer Network. BJU Int. 103, 747–752 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Abrahams NA, Moran C, Reyes AO, Siefker-Radtke A & Ayala AG Small cell carcinoma of the bladder: a contemporary clinicopathological study of 51 cases. Histopathology 46, 57–63 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Lynch SP et al. Neoadjuvant chemotherapy in small cell urothelial cancer improves pathologic downstaging and long-term outcomes: results from a retrospective study at the MD Anderson Cancer Center. Eur. Urol. 64, 307–313 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siefker-Radtke AO et al. Evidence supporting preoperative chemotherapy for small cell carcinoma of the bladder: a retrospective review of the M. D. Anderson cancer experience. J. Urol. 172, 481–484 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Thota S, Kistangari G, Daw H & Spiro T A clinical review of small-cell carcinoma of the urinary bladder. Clin. Genitourin. Cancer 11, 73–77 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Geynisman DM et al. Advanced small cell carcinoma of the bladder: clinical characteristics, treatment patterns and outcomes in 960 patients and comparison with urothelial carcinoma. Cancer Med. 5, 192–199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bex A et al. Small cell carcinoma of bladder: a single-center prospective study of 25 cases treated in analogy to small cell lung cancer. Urology 65, 295–299 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Choong NWW, Quevedo JF & Kaur JS Small cell carcinoma of the urinary bladder. The Mayo Clinic experience. Cancer 103, 1172–1178 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Pasquier D et al. Small Cell Carcinoma of the Urinary Bladder: A Retrospective, Multicenter Rare Cancer Network Study of 107 Patients. Int. J. Radiat. Oncol. Biol. Phys. 92, 904–910 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Mattes MD, Kan C-C, Dalbagni G, Zelefsky MJ & Kollmeier MA External beam radiation therapy for small cell carcinoma of the urinary bladder. Pract. Radiat. Oncol. 5, e17–22 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Patel SG et al. Locoregional small cell carcinoma of the bladder: clinical characteristics and treatment patterns. J. Urol. 191, 329–334 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Siefker-Radtke AO et al. Phase II clinical trial of neoadjuvant alternating doublet chemotherapy with ifosfamide/doxorubicin and etoposide/cisplatin in small-cell urothelial cancer. J Clin Oncol. 27, 2592–2597 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meijer RP et al. Local control rate and prognosis after sequential chemoradiation for small cell carcinoma of the bladder. Int. J. Urol. Off. J. Jpn. Urol. Assoc. 20, 778–784 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Kollmeier MA COUNTERPOINT: Is Cystectomy Needed for Small-Cell Bladder Cancer? Oncol. Williston Park N 29, 645, 648–649 (2015). [PubMed] [Google Scholar]
- 19.Raghavan D POINT: Is Cystectomy Needed for Small-Cell Bladder Cancer? Oncol. Williston Park N 29, 645–647 (2015). [PubMed] [Google Scholar]
- 20.Kouba EJ & Cheng L Understanding the Genetic Landscape of Small Cell Carcinoma of the Urinary Bladder and Implications for Diagnosis, Prognosis, and Treatment: A Review. JAMA Oncol. (2017). doi: 10.1001/jamaoncol.2016.7013 [DOI] [PubMed] [Google Scholar]
- 21.Teo M et al. Small cell carcinoma of the bladder (SCCB): Clinical, histopathologic, and genomic predictors of clinical outcomes. J Clin Oncol. 35, 294–294 (2017). [Google Scholar]
- 22.Ladi E et al. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J. Cell Biol. 170, 983–992 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunders LR et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci. Transl. Med. 7, 302ra136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dylla SJ Toppling high-grade pulmonary neuroendocrine tumors with a DLL3-targeted trojan horse. Mol. Cell. Oncol. 3, e1101515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudin CM et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 18, 42–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen N et al. Transcriptional gene expression profiling of small cell lung cancer cells. Cancer Res. 63, 1943–1953 (2003). [PubMed] [Google Scholar]
- 27.Peifer M et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 44, 1104–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George J et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourzac KM et al. A high-density quantitative nuclease protection microarray platform for high throughput analysis of gene expression. J. Biotechnol. 154, 68–75 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Roberts RA et al. Quantitative nuclease protection assay in paraffin-embedded tissue replicates prognostic microarray gene expression in diffuse large-B-cell lymphoma. Lab. Investig. J. Tech. Methods Pathol. 87, 979–997 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Pharmaceutical Statistics: MBSW 39, Muncie, Indiana, USA, May 16–18, 2016. (Springer International Publishing, 2018). [Google Scholar]
- 32.VENTANA PD- L1 (SP263) Assay Available at: https://www.accessdata.fda.gov/cdrh_docs/pdf16/p160046c.pdf (Accessed September 3, 2018).
- 33.VENTANA PD- L1 (SP142) Assay Available at: https://www.accessdata.fda.gov/cdrh_docs/pdf16/P160002c.pdf (Accessed September 3, 2018).
- 34.Kim KH & Roberts CWM Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato T et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci. Rep. 3, 1911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coe BP et al. Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer. PloS One 8, e71670 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang MT et al. Small cell carcinomas of the bladder and lung are characterized by a convergent but distinct pathogenesis. Clin Cancer Res. (2017). doi: 10.1158/1078-0432.CCR-17-2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faltas BM et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat. Genet. 48, 1490–1499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunders LR et al. Expression of DLL3 in metastatic melanoma, glioblastoma and high-grade extrapulmonary neuroendocrine carcinomas as potential indications for rovalpituzumab tesirine (Rova-T; SC16LD6.5), a delta-like protein 3 (DLL3)-targeted antibody drug conjugate (ADC). Am. Assoc. Cancer Res. AACR Annu. Meet. Proc. Abstract 3093, (2017). [Google Scholar]
- 41.Rovalpituzumab Tesirine in Delta-Like Protein 3-Expressing Advanced Solid Tumors - Full Text View - ClinicalTrials.gov. Available at: https://clinicaltrials.gov/ct2/show/NCT02709889?term=DLL3&rank=4. (Accessed: 16th March 2017)
- 42.Sharma SK et al. Fc-Mediated Anomalous Biodistribution of Therapeutic Antibodies in Immunodeficient Mouse Models. Cancer Res. 78, 1820–1832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuruoka K et al. PD-L1 expression in neuroendocrine tumors of the lung. Lung Cancer Amst. Neth. 108, 115–120 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


