FIGURE 5.
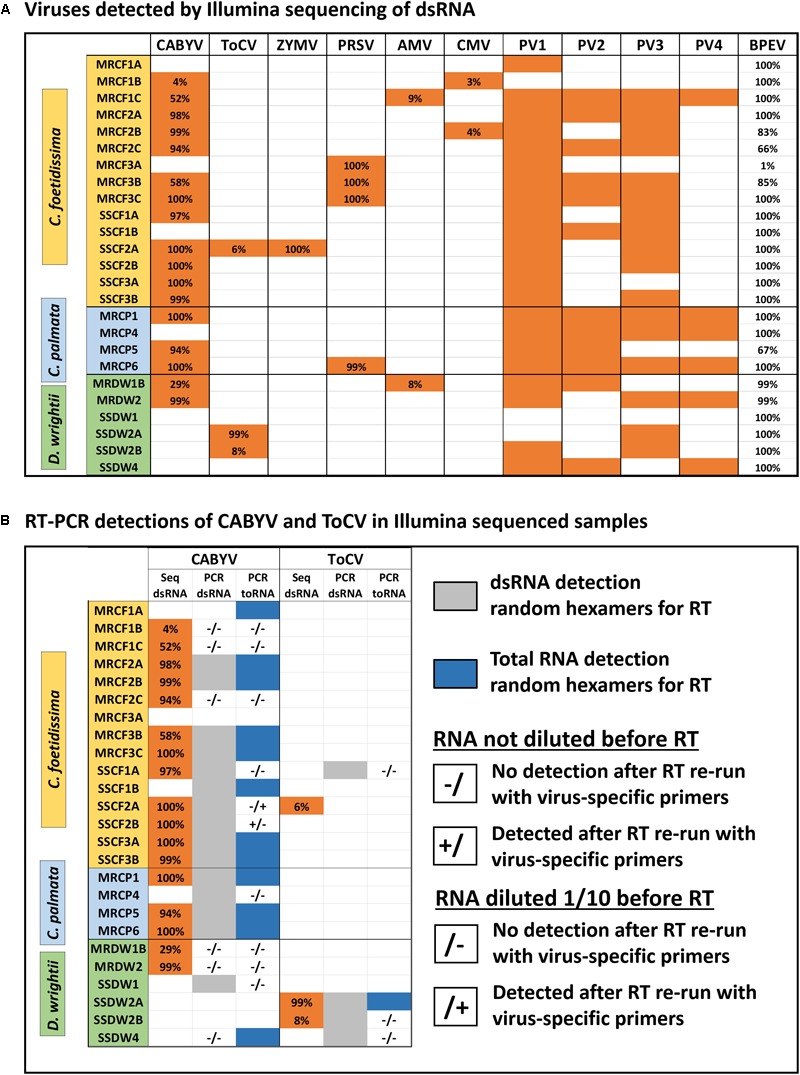
Viromes of individual plants (A) and summary of detections by different diagnostic methods for select crop-associated viruses (B). Sample codes within each host plant section indicate species (CF, CP, or DW), site of collection (MR, Motte Rimrock; SS, Shipley-Skinner), numbers (1, 2, and 3) indicate populations within a site, and letters (A, B, and C) indicate individuals within each population. Viruses are as follows, from left to right: Cucurbit aphid-borne yellows virus, Tomato chlorosis virus, Zucchini yellow mosaic virus, Papaya ringspot virus, Alfalfa mosaic virus, and Cucumber mosaic virus. PV1–4 refer to novel viruses related to members of the family Partitiviridae. The last column shows the coverage of the internal control (Bell pepper endornavirus) for each sample. Results are from assemblies constructed after subtraction of host genomes (see the section “Materials and Methods”). We found no effect of host subtraction on the identity and number of viruses detected (see the section “Evaluating Detection Biases in Bioinformatic Workflows and Downstream Applications” in the Discussion).
