Abstract
Monocarboxylate transporters (MCTs) mediate the proton-coupled exchange of high-energy metabolites, including lactate and pyruvate, between cells and tissues. The transport activity of MCT1, MCT2, and MCT4 can be facilitated by the extracellular carbonic anhydrase IV (CAIV) via a noncatalytic mechanism. Combining physiological measurements in HEK-293 cells and Xenopus oocytes with pulldown experiments, we analyzed the direct interaction between CAIV and the two MCT chaperones basigin (CD147) and embigin (GP70). Our results show that facilitation of MCT transport activity requires direct binding of CAIV to the transporters chaperones. We found that this binding is mediated by the highly conserved His-88 residue in CAIV, which is also the central residue of the enzyme's intramolecular proton shuttle, and a charged amino acid residue in the Ig1 domain of the chaperone. Although the position of the CAIV-binding site in the chaperone was conserved, the amino acid residue itself varied among different species. In human CD147, binding of CAIV was mediated by the negatively charged Glu-73 and in rat CD147 by the positively charged Lys-73. In rat GP70, we identified the positively charged Arg-130 as the binding site. Further analysis of the CAIV-binding site revealed that the His-88 in CAIV can either act as H donor or H acceptor for the hydrogen bond, depending on the charge of the binding residue in the chaperone. Our results suggest that the CAIV-mediated increase in MCT transport activity requires direct binding between CAIV–His-88 and a charged amino acid in the extracellular domain of the transporter's chaperone.
Keywords: electrophysiology, energy metabolism, ion-sensitive electrode, lactic acid, membrane transport, pH regulation, protein complex, protein expression, proton transport, Xenopus
Introduction
The SLC16 gene family of monocarboxylate transporters (MCT)3 comprises 14 isoforms, the first four of which (MCT1–4) carry high-energy metabolites, including lactate, pyruvate, and ketone bodies together with H+ in a 1:1 stoichiometry across the plasma membrane (1–4). MCT1, which is found in nearly every tissue, has a Km of 3–5 mm for l-lactate (1, 2). MCT2, which is primarily found in liver, kidney, testis, and brain (5–7), has the highest affinity for l-lactate among all MCTs with a Km of about 0.7 mm (8). In liver and kidney, MCT2 facilitates the uptake of lactate, which is then used for glyconeogenesis (3). In the brain, MCT2 is expressed in neurons, where it facilitates the import of lactate, which is released by astrocytes and vascular endothelial cells via MCT1 and MCT4 (9–12). Expression of MCT3 is restricted to retinal pigment epithelium and choroid plexus epithelia, where it primarily serves as a lactate exporter (13–15). MCT3 transports l-lactate with a Km of about 6 mm (16). MCT4 is a low-affinity, high-capacity carrier with a Km of 20–35 mm for l-lactate (17), which primarily acts as a lactate exporter in glycolytic cells and tissues like astrocytes, skeletal muscle, and (hypoxic) tumor cells (3, 4, 18, 19). All MCTs have a 12 transmembrane-helix structure, with both the C and N termini located intracellularly (3, 4). Trafficking, but also regulation of transport activity of MCT1–4, is mediated by ancillary proteins. MCT1, MCT3, and MCT4 are associated with CD147 (also termed basigin, EMMPRIN, OX-47, HAb18G, or HT7), whereas surface expression of MCT2 requires GP70 (also termed embigin) (20–23). The ancillary proteins remain tightly associated with the transporter in the membrane, which is essential to maintain transporter activity (22, 24). Furthermore, the chaperones modulate the sensitivity of MCTs to different inhibitors (22, 25, 26). Both chaperones are glycoproteins that belong to the immunoglobulin superfamily. CD147 is a multifunctional protein, which mediates various molecular events crucial to many biological functions. CD147 is involved in spermatogenesis and fertilization (27, 28), as well as neural network formation and development (29, 30). It plays a central role in the inflammatory response pathway and is used as a receptor by viruses, including the human immunodeficiency virus (HIV) (31, 32). Furthermore, CD147 is highly expressed on the surface of tumor cells, where it stimulates adjacent fibroblasts and tumor cells to produce matrix metalloproteinases, leading to cancer progression (33).
CD147 comprises a transmembrane domain, a short intracellular C-terminal tail, and two or three extracellular immunoglobulin (Ig)-like domains (34–36). The general isoform, CD147 Ig1–Ig2 (also termed basigin or basigin-2), features two Ig-like domains, whereas the retinal-specific isoform CD147 Ig0–Ig1–Ig2 (basigin-1) contains an additional Ig-like domain distal to the transmembrane domain (35, 36). The Ig1–Ig2 domain of CD147 Ig1–Ig2 (used in this study) has been crystallized, and the structure was solved at 2.8 Å resolution (37). The N-terminal Ig1 domain (D1, residues 22–101) strongly resembles the C2 set fold, whereas the C-terminal Ig2 domain (D2, residues 107–205) shows strong homology to the Ig variable domain (V set) (37). Both Ig domains are connected by a flexible, five amino acids long linker (37).
Like CD147, GP70 has a short intracellular C-terminal tail, a single transmembrane domain, and a large extracellular N terminus, containing two immunoglobulin-like domains (Ig1, residues 88–145; Ig2, residues 181–239) (26, 38). GP70 has been reported to be a modulator of neuromuscular junction formation and enhancer of neuromuscular adhesion. Furthermore, it facilitates integrin-mediated cell–to–substratum adhesion and cell–to–extracellular matrix interactions (22, 39, 40). Moreover, recent studies demonstrated that GP70 is involved in the progression of breast and pancreatic cancer (40, 41).
Xenopus oocytes, used in this study, endogenously express CD147, which can mediate integration of heterologously expressed MCT1 and MCT4 into the oocyte plasma membrane, whereas GP70, the preferred ancillary protein of MCT2, is not endogenously expressed in these cells (25).
Mammalian carbonic anhydrases (CA) included in the α-class of CAs, of which 16 isoforms are identified, catalyze the reversible hydration of CO2 to HCO3− and H+ (42). CAIV, used in this study, is anchored to the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor, with the catalytic domain facing the extracellular space (43). CAIV is expressed in various organs and tissues, including heart, lung, kidney, brain, retina, and erythrocytes (44).
CAIV enhances transport activity of several other acid/base transporters, including the Cl−/HCO3− exchangers AE1–3 (45, 46), the Na+/HCO3− cotransporter NBCe1 (47), and the Na+/H+ exchanger NHE1 (48). In all cases, interaction between transporter and CAIV requires both CA catalytic activity and the physical interaction between the enzyme and an extracellular domain of the respective transporter.
CAIV further facilitates transport activity of the monocarboxylate transporters MCT1, MCT2, and MCT4 (49–51). In contrast to the “transport metabolons” described before, augmentation of MCT activity by CAIV did not depend on the catalytic activity of the enzyme, because neither application of the CA inhibitor 6-ethoxy-2-benzothiazolesulfonamide nor coexpression of the catalytically inactive mutant CAIV-V165Y had any effect on the CAIV-mediated increase in MCT transport activity (49). A similar type of interaction was also observed between intracellular CAII and MCT1/MCT4 (52–55). Because CAII catalytic activity is not required to facilitate MCT transport function, it was hypothesized that CAII might utilize parts of its intramolecular proton pathway to function as a proton antenna for the transporter (55–57). Protonable residues with overlapping Coulomb cages could form proton-attractive domains and could share a proton at a very fast rate, exceeding the upper limit of diffusion-controlled reactions (58, 59). When these residues are located in proteins or lipid headgroups at the plasma membrane, they can collect protons from the solution and direct them to the entrance of a proton–transfer pathway of membrane-anchored proteins, a phenomenon termed “proton-collecting antenna” (58, 60). The need for such a proton antenna is based on the observation that H+ cotransporters, such as MCTs, extract H+ from the surrounding area at rates well above the capacity for simple diffusion to be replenished from their immediate vicinity. Therefore, the transporter must exchange H+ with protonable sites at the plasma membrane, which could function as proton collectors for the transporter (61). Proton transfer between transporter and enzyme requires close proximity between the two proteins. CAII binds MCT1 and MCT4 via a cluster of three glutamic acid residues within the transporters' C-terminal tails (MCT1, 489EEE (62), and MCT4, 431EEE (63)). In both cases, the two outer glutamic acids form hydrogen bonds with the histidine at position 64 in CAII (57).
CAIV-mediated facilitation of MCT transport activity required the enzyme to be localized on the extracellular surface of the oocyte (49). Furthermore, the interaction of MCT2 and CAIV was only detectable when MCT2 was coexpressed with its ancillary protein GP70. From this, it was hypothesized that MCT2 and CAIV may not interact directly with each other, but may require GP70 as a mediator, possibly by allowing binding of CAIV to one of the Ig domains.
In this study, we investigated the interaction between CAIV and the two MCT chaperones CD147 and GP70. Our results show that facilitation of MCT transport activity requires direct binding of CAIV to the chaperone. Binding between the proteins is thereby mediated by the highly conserved CAIV–His-88, which is also the central residue of the enzyme's intramolecular proton shuttle, and a charged amino acid residue in the Ig1 domain of CD147 and GP70, respectively.
Results
Facilitation of MCT transport activity by CAIV requires the Glu-73 in the Ig1 domain of hCD147
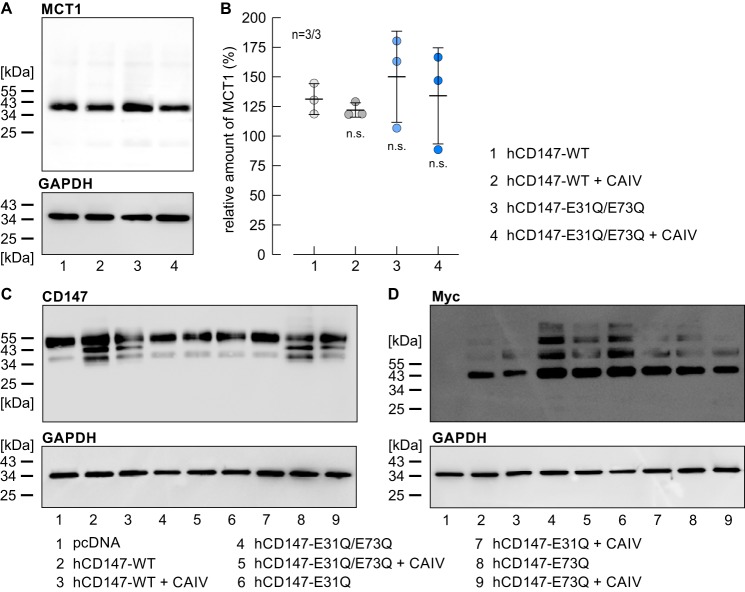
We have previously shown that CAIV facilitates transport activity of MCT1, MCT2, and MCT4 by a mechanism that is independent from the enzyme's catalytic activity, presumably by functioning as a proton antenna for the transporter (49–51). Proton transfer between MCT and CAIV would require close proximity between the proteins, which could be achieved by direct binding. Intracellular CAII binds to MCT1 and MCT4 via a cluster of three glutamic acid residues within the transporter's C-terminal tail (MCT1, 489EEE (62), and MCT4, 431EEE (63)). In both cases, the two outer glutamic acids form hydrogen bonds with the histidine at position 64 in CAII (57). To investigate whether CAIV-mediated facilitation of MCT transport activity requires binding of CAIV to the transporter's chaperon, we first searched for a possible CAIV-binding site in the human isoform of CD147, the chaperone of MCT1 and MCT4. A structural model of the human MCT1–CD147 protein complex (26) revealed glutamic acid residues with a similar arrangement to MCT1–489EEE and MCT4–431EEE. Subsequent molecular docking of CAIV (PDB code 5JN9) to hCD147 (PDB code 3B5H) identified the two glutamic acid residues, Glu-31 and Glu-73 in the Ig1 domain of hCD147 as potential interaction partners for CAIV–His-88 (the analogous residue to CAII–His-64) (Fig. 1A). To investigate whether Glu-31 and Glu-73 are involved in the functional interaction between MCT–CD147 and CAIV, we coexpressed hCD147–WT or a mutant of hCD147, in which either Glu-31 or Glu-73, or both residues, were mutated to glutamine, together with CAIV in human embryonic kidney (HEK) 293 cells. Because HEK-293 cells express robust levels of endogenous MCT1 (64), no additional MCTs were expressed. MCT transport activity was determined by measuring the rate of change in intracellular H+ concentration (Δ[H+]/Δt) during application of 3 and 10 mm lactate in HEK-293 cells, transfected with hCD147 alone or together with CAIV (Fig. 1B). Coexpression of hCD147–WT with CAIV resulted in a significant increase in Δ[H+]/Δt, indicating CAIV-mediated facilitation of MCT transport activity in HEK-293 cells (Fig. 1C). Coexpression of hCD147–E31Q with CAIV resulted in a similar increase in MCT transport activity. However, no increase in MCT transport activity could be observed when hCD147–E73Q or the double mutant hCD147–E31Q/E73Q were coexpressed with CAIV (Fig. 1, B and C). These results indicate that CAIV-mediated facilitation of MCT transport activity requires Glu-73 in the Ig1 domain of hCD147.
Figure 1.
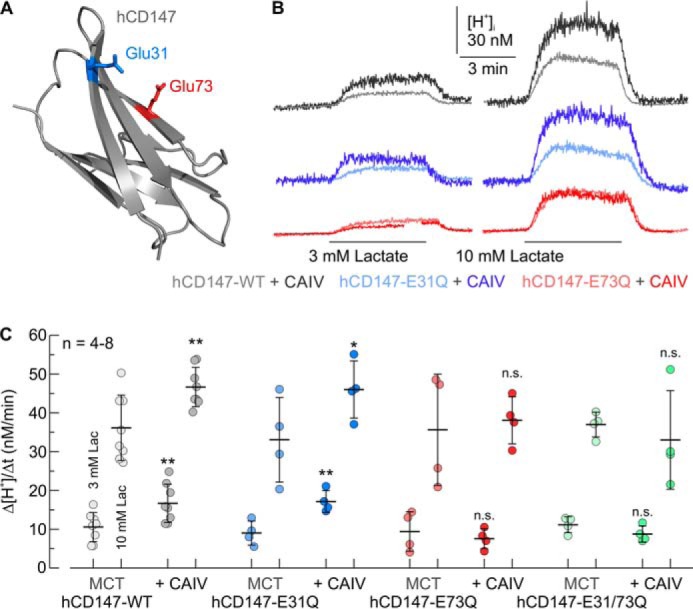
Facilitation of MCT transport activity by CAIV in HEK-293 cells requires Glu-73 in the Ig1 domain of the transporter's chaperone CD147. A, structure of the Ig1 domain of human CD147 (PDB code 3B5H (37)). Glu-31 and Glu-73 are labeled in blue and red, respectively. B, original recordings of the change in intracellular H+ concentration during application of lactate in HEK-293 cells, transfected with hCD147–WT (gray traces), hCD147–E31Q (blue traces), and hCD147–E73Q (red traces), respectively. Cells were either transfected with hCD147 alone (light traces) or cotransfected with hCD147 and human CAIV (dark traces). C, rate of change in intracellular H+ concentration (Δ[H+]/Δt) during application of lactate in HEK-293 cells, transfected with hCD147–WT (gray dots), hCD147–E31Q (blue dots), hCD147–E73Q (red dots), and the double-mutant hCD147–E31Q/E73Q (green dots). Cells were either transfected with hCD147 alone (light dots) or cotransfected with hCD147 and human CAIV (dark dots). The significance indicators above the dots with CAIV (dark dots) refer to the corresponding dots without CAIV (light dots of same color).
To check whether expression levels of endogenous MCT1 in HEK-293 cells were influenced by transfection of the cells with hCD147 and CAIV, we performed a Western blot analysis of MCT1 in HEK-293 cells, transfected with hCD147–WT or hCD147–E31Q/E73Q either alone or together with CAIV (Fig. 2A). Quantification of the signal for MCT1 showed that expression of endogenous MCT1 was not significantly changed by transfection with either one of the constructs (Fig. 2B). Expression levels of whole CD147 (endogenous protein and transfected hCD147) as well as expression levels of the transfected Myc-tagged hCD147 were assessed by Western blottings against CD147 (Fig. 2C) and Myc tag (Fig. 2D). The blots against CD147 showed a band at 55 kDa for all samples and some lower molecular weight bands, which might indicate protein degradation. The blots against Myc-tagged CD147 presented bands at 40 kDa for cells transfected with the hCD147 variants with and without CAIV, but not for the cells transfected with the empty vector pcDNA3 (Fig. 2D, lane 1). The extra bands above the expected molecular weight could be due to glycosylation of the protein or oligomerization with degradation products.
Figure 2.
Expression of MCT1 and CD147 in HEK-293 cells. A, representative Western blotting against MCT1 (upper blot) and GAPDH (lower blot) as loading control, from HEK-293 cells transfected with human CD147–WT (lane 1), hCD147–WT, and CAIV (lane 2), the mutant hCD147–E31Q/E73Q (lane 3), and hCD147–E31Q/E73Q and CAIV (lane 4), respectively. B, quantification of the relative protein level of MCT1 (as normalized to the signal of GAPDH in the same lane) in HEK-293 cells, transfected with hCD147–WT (lane 1), hCD147–WT + CAIV (lane 2), hCD147–E31Q/E73Q (lane 3), and hCD147–E31Q/E73Q + CAIV (lane 4), respectively. The significance indicators refer to the values of HEK-293 cells, transfected with hCD147–WT. n is given as the number of Western blots/number of batches of cells. C, representative Western blotting against CD147 (upper blot) and GAPDH (lower blot) as loading control, from HEK-293 cells, transfected with the empty vector pcDNA3 (lane 1), hCD147–WT or a mutant of CD147 (lanes 2, 4, 6, and 8), or cotransfected with hCD147 or a mutant of CD147 and CAIV (lanes 3, 5, 7, and 9). D, representative Western blotting against the Myc tag of the transfected CD147 (upper blot) and GAPDH (lower blot) as loading control, from HEK-293 cells, transfected with the empty vector pcDNA3.1 (lane 1), hCD147–WT or a mutant of hCD147 (lanes 2, 4, 6, and 8), or cotransfected with hCD147 or a mutant of hCD147 and CAIV (lanes 3, 5, 7, and 9).
To test whether Glu-73 in hCD147 functions as the binding site for CAIV, we performed pulldown experiments with CAIV and GST fusion proteins of the Ig1 domain of hCD147–WT, hCD147–E31Q, and hCD147–E73Q, respectively (Fig. 3A). GST alone was used as negative control. Pulldown of CAIV with GST-hCD147–WT resulted in a robust signal for CAIV, indicating direct binding of CAIV to the Ig1 domain of hCD147 (Fig. 3, A and B). Mutation of Glu-73 to Gln resulted in a significant reduction in the signal, which indicates loss of binding between hCD147–E73Q and CAIV, whereas the mutation E31Q had no significant influence on binding (Fig. 3, A and B). From this, we conclude that binding of CAIV is mediated by Glu-73 in the Ig1 domain of hCD147.
Figure 3.
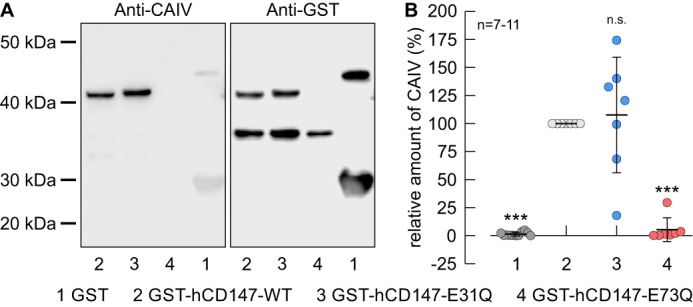
Binding of CAIV to hCD147 requires the Glu-73 in the Ig1 domain of hCD147. A, representative Western blots of CAIV (left blot) and GST (right blot), respectively. CAIV was pulled down with GST (lane 1), a GST fusion protein of the Ig1 domain of hCD147–WT (lane 2), a GST fusion protein of Ig1 domain of the hCD147 mutant E31Q (lane 3), and a GST fusion protein of Ig1 domain of the hCD147 mutant E73Q (lane 4). B, relative intensity of the fluorescent signal of CAIV. For every blot, the signals for CAIV were normalized to the corresponding signals for GST–hCD147–WT. Each individual signal for CAIV was normalized to the intensity of the signal for GST in the same lane. The significance indicators above the dots refer to GST–hCD147–WT.
Interaction between CAIV and rat MCT1 requires Lys-73 in rCD147
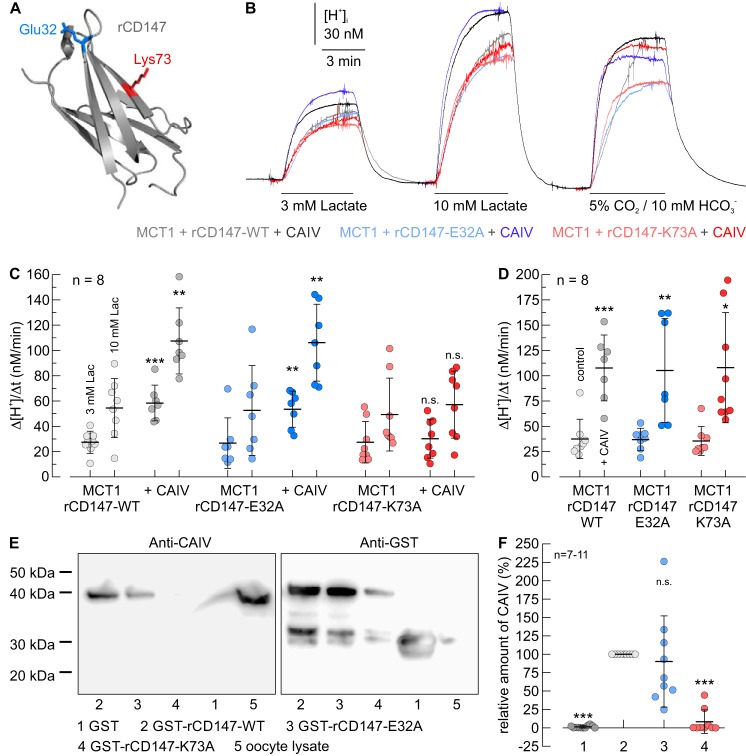
Interestingly, Glu-73, which mediates binding to CAIV in human CD147, is not conserved among the mammalian species. Although CD147 of human and rabbit feature glutamic acid at position 73, CD147 from rat and mouse feature a lysine at this position (Figs. 4 and 5A). To investigate whether functional interaction between CAIV and rat MCT1 requires the Lys-73 in rat CD147, we coexpressed rMCT1 together with rCD147–WT, rCD147–E32A, or rCD147–K73A in Xenopus oocytes. Cells either expressed rMCT1 + rCD147 alone or together with CAIV. rMCT1 transport activity was determined by measuring the rate of change in intracellular H+ concentration (Δ[H+]/Δt) during application of 3 and 10 mm lactate (Fig. 5, B and C). In the absence of CAIV, no significant differences in Δ[H+]/Δt could be detected when rMCT1 was coexpressed with rCD147–WT, rCD147–E32A, or rCD147–K73A, indicating equal expression of all the three protein complexes (Fig. 5C). Coexpression of rMCT1 + rCD147–WT and MCT1 + rCD147–E32A with CAIV resulted in an ∼2-fold increase in rMCT1 transport activity. However, coexpression of CAIV with rMCT1 + rCD147–K73A did not increase transport function (Fig. 5C). Catalytic activity of CAIV was controlled in each oocyte by measuring Δ[H+]/Δt during application of 5% CO2, 10 mm HCO3− at the end of the experiment (Fig. 5B). In all three batches of oocytes, expression of CAIV increased the rate of CO2-induced acidification by a factor of ∼3, with no significant difference between the batches, indicating equal expression of CAIV protein in all three batches of oocytes (Fig. 5D). The influence of CAIV on the expression level of rMCT1 was already investigated in a previous study, which demonstrated that coexpression of CAIV does not result in an increase in rMCT1 protein level (50).
Figure 4.

Alignment of the protein sequence, surrounding the CAIV-binding site of CD147 from human (BAA08109.1), rabbit (NP_001075843.1), rat (AAH59145.1), and mouse (BAA00486.1). The CAIV-binding site at position 73 is marked in red.
Figure 5.
Functional interaction between rat MCT1 and CAIV in Xenopus oocytes requires direct binding of CAIV to Lys-73 in the Ig1 domain of rCD147. A, homology model of the Ig1 domain of rat CD147 (based on human CD147, PDB code 3B5H (37)). Glu-32 and Lys-73 are labeled in blue and red, respectively. B, original recordings of the change in intracellular H+ concentration during application of lactate and CO2/HCO3− in Xenopus oocytes, expressing rMCT1 together with rCD147–WT (gray traces), rCD147–E32A (blue traces), and rCD147–K73A (red traces), respectively. Cells were either expressing rMCT1 + rCD147 alone (light traces) or rMCT1 + rCD147 and human CAIV (dark traces). C and D, rate of change in intracellular H+ concentration (Δ[H+]/Δt) during application of lactate (C) and 5% CO2, 10 mm HCO3− (D) in Xenopus oocytes, expressing rMCT1 and rCD147–WT (gray dots), rCD147–E32A (blue dots), and rCD147–K73A (red dots), respectively. Cells were either expressing rMCT1 + rCD147 alone (light dots) or rMCT1 + rCD147 and CAIV (dark dots). The significance indicators above the dots with CAIV (dark dots) refer to the corresponding dots without CAIV (light dots of same color). E, representative Western blots of CAIV (left blot) and GST (right blot), respectively. CAIV was pulled down with GST (lane 1), a GST fusion protein of the Ig1 domain of rCD147–WT (lane 2), a GST fusion protein of Ig1 domain of rCD147–E32A (lane 3), and a GST fusion protein of Ig1 domain of rCD147–K73A (lane 4). Lysate of CAIV-expressing oocytes (lane 5) was added as positive control. F, relative intensity of the fluorescent signal of CAIV. For every blot, the signals for CAIV were normalized to the corresponding signals for GST–rCD147–WT. Each individual signal for CAIV was normalized to the intensity of the signal for GST in the same lane. The significance indicators above the dots refer to GST–rCD147–WT.
To test whether Lys-73 in rCD147 also functions as a binding site for CAIV, we performed pulldown experiments with CAIV and GST fusion proteins of the Ig1 domain of rCD147–WT, rCD147–E32A, and rCD147–K73A, respectively (Fig. 5E). GST alone was used as negative control. Pulldown of CAIV with GST–rCD147–WT resulted in a robust signal for CAIV, indicating direct binding of CAIV to the Ig1 domain of rCD147 (Fig. 5, E and F). Mutation of Lys-73 to Ala resulted in an almost complete loss of the signal, which indicates that binding between rCD147–K73A and CAIV failed, whereas the mutation E32A had no significant influence on binding (Fig. 5, E and F). Taken together, these results suggest that the functional interaction between rMCT1/rCD147 and CAIV requires direct binding of the enzyme to Lys-73 in the Ig1 domain of rCD147.
Interaction between CAIV and rat MCT2 requires Arg-130 in rGP70
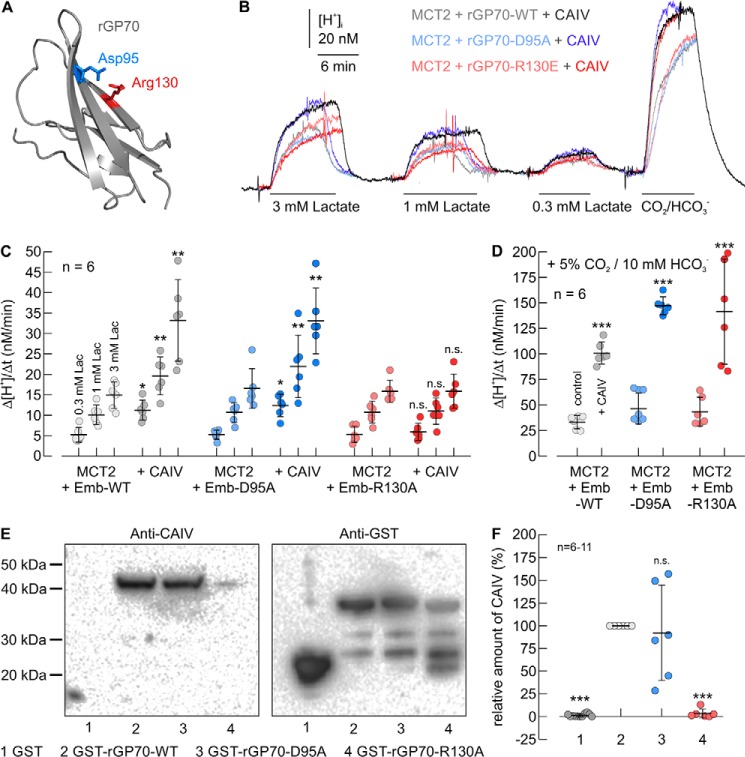
Although MCT1 and MCT4 are associated with CD147, surface expression and transport activity of MCT2 are facilitated by GP70. To elucidate a possible CAIV-binding site of GP70, we created a homology model of rGP70, based on the structure of hCD147 (Fig. 6A). Thereby, we identified the two amino acids Asp-95 and Arg-130 in the Ig1 domain of rGP70 as corresponding to Glu-31 and Glu-73 in hCD147 for binding CAIV. Both residues are conserved between the human and rat isoform of GP70.
Figure 6.
Functional interaction between rat MCT2 and CAIV in Xenopus oocytes requires direct binding of CAIV to Arg-130 in the Ig1 domain of rGP70. A, homology structure of the Ig1 domain of rat GP70 (based on human CD147, PDB code 3B5H (37)). Glu-95 and Arg-130 are labeled in blue and red, respectively. B, original recordings of the change in intracellular H+ concentration during application of lactate and CO2/HCO3− in Xenopus oocytes, expressing rMCT2 together with rGP70–WT (gray traces), rGP70–D95A (blue traces), and rGP70–R130A (red traces), respectively. Cells were either expressing rMCT2 + rGP70 alone (light traces) or rMCT2 + rGP70 and human CAIV (dark traces). C and D, rate of change in intracellular H+ concentration (Δ[H+]/Δt) during application of lactate (C) and 5% CO2, 10 mm HCO3− (D) in Xenopus oocytes, expressing rMCT2 and rGP70–WT (gray dots), rGP70–D95A (blue dots), and rGP70–R130A (red dots), respectively. Cells were either expressing rMCT2 + rGP70 alone (light dots) or rMCT1 + rGP70 and human CAIV (dark dots). The significance indicators above the dots with CAIV (dark dots) refer to the corresponding dots without CAIV (light dots of same color). E, representative Western blots of CAIV (left blot) and GST (right blot), respectively. CAIV was pulled down with GST (lane 1), a GST fusion protein of the Ig1 domain of rGP70–WT (lane 2), a GST fusion protein of Ig1 domain of rGP70–D95A (lane 3), and a GST fusion protein of Ig1 domain of rGP70–R130A (lane 4). F, relative intensity of the fluorescent signal of CAIV. For every blot, the signals for CAIV were normalized to the corresponding signals for GST–rGP70–WT. Each individual signal for CAIV was normalized to the intensity of the signal for GST in the same lane. The significance indicators above the dots refer to GST–rGP70–WT.
To investigate whether Asp-95 or Arg-130 is required for the functional interaction between MCT2 + GP70 and CAIV, we coexpressed rMCT2 together with rGP70–WT, rGP70–D95A, or rGP70–R130A in Xenopus oocytes. Cells either expressed rMCT2 + rGP70 alone or together with CAIV. rMCT2 transport activity was determined by measuring Δ[H+]/Δt during application of 0.3, 1, and 3 mm lactate (Fig. 6, B and C). In the absence of CAIV no significant differences in Δ[H+]/Δt could be detected when rMCT2 was coexpressed with rGP70–WT, rGP70–D95A, or rGP70–R130A, respectively, indicating equal expression of all the three protein complexes (Fig. 6C). Coexpression of rMCT2 + rGP70–WT and rMCT2 + rGP70–D95A with CAIV resulted in an ∼2-fold increase in rMCT2 transport activity. However, coexpression of CAIV with rMCT2 + rGP70–R130A did not increase rMCT2 transport function (Fig. 6C). Catalytic activity of CAIV was again tested in each oocyte by measuring Δ[H+]/Δt during application of 5% CO2, 10 mm HCO3− (Fig. 6B). In all three batches of oocytes, expression of CAIV increased the rate of CO2-induced acidification by a factor of ∼3, with no significant difference between the batches (Fig. 6D).
To test whether Arg-130 in rGP70 also functions as a binding site for CAIV, we performed pulldown experiments with CAIV and GST fusion proteins of the Ig1 domain of rGP70–WT, rGP70–D95A, and rGP70–R130A, respectively (Fig. 6E). GST alone was used as negative control. Pulldown of CAIV with GST–rGP70–WT resulted in a robust signal for CAIV, indicating direct binding of CAIV to the Ig1 domain of rGP70 (Fig. 6, E and F). Mutation of Arg-130 to Ala resulted in an almost complete loss of the signal, which indicates loss of binding between rGP70–R130A and CAIV, whereas the mutation D96A had no significant influence on binding (Fig. 6, E and F). Taken together, these results indicate that the functional interaction between rMCT2/rGP70 and CAIV requires direct binding of the enzyme to Arg-130 in the Ig1 domain of rGP70.
Binding of CAIV to GP70 is mediated by His-88 in CAIV
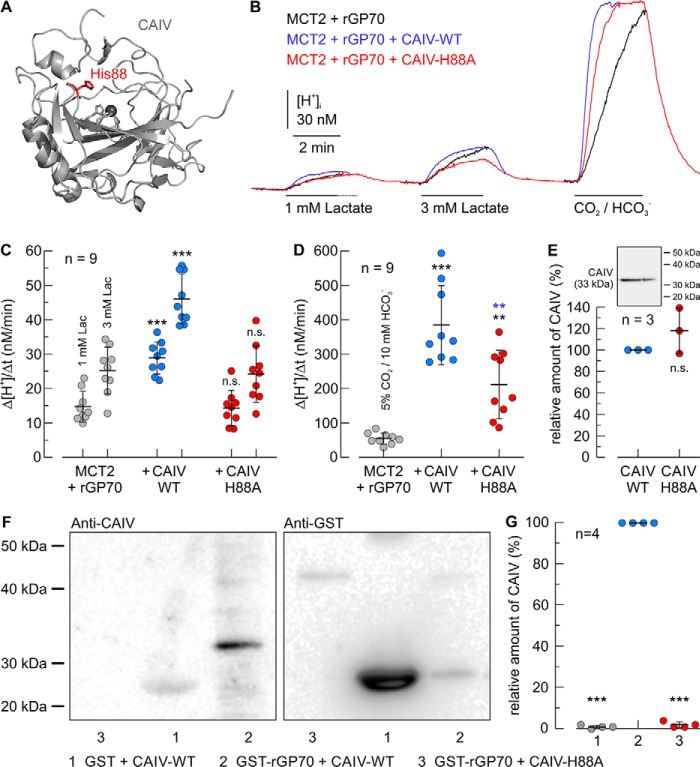
We have shown previously that binding of intracellular CAII to the C-terminal tail of MCT1 and MCT4 is mediated by the His-64 in CAII (57, 62, 63). The analog residue to CAII–His-64 in CAIV is His-88 (sometimes also termed CAIV–His-64, in analogy to the His-64 in CAII). Because His-88 also serves as the central residue of the enzyme's intramolecular proton shuttle, and is fundamental for CAIV enzymatic activity, it is conserved along all mammalian species. The position of His-88 is depicted in Fig. 7A. To investigate the role of CAIV–His-88 in the interaction with the MCT–chaperone complex, we coexpressed rMCT2 and rGP70 with CAIV–WT or CAIV–H88A in Xenopus oocytes. rMCT2 transport activity was determined by measuring Δ[H+]/Δt during application of 1 and 3 mm lactate (Fig. 7, B and C). Although transport activity of rMCT2 was again increased ∼2-fold in the presence of CAIV–WT, CAIV–H88A failed to facilitate rMCT2 transport activity. Catalytic activity of CAIV was determined by application of 5% CO2, 10 mm HCO3− (Fig. 7, B and D). Coexpression of rMCT2/rGP70 with CAIV–WT increased the rate of CO2-induced acidification by a factor of ∼7, whereas coexpression with CAIV–H88A increased Δ[H+]/Δt only by a factor of ∼4, indicating a reduction in CAIV–H88A catalytic activity. Western blot analysis, however, indicated no significant difference in the expression levels of CAIV–WT and CAIV–H88A (Fig. 7E).
Figure 7.
Functional interaction between rat MCT2 and CAIV in Xenopus oocytes requires direct binding of rGP70 to His-88 in CAIV. A, structure of human CAIV (PDB code 5JN9 (79). The amino acid residue His-88 is labeled in red. B, original recordings of the change in intracellular H+ concentration during application of lactate and CO2/HCO3− in Xenopus oocytes, expressing rMCT2 + rGP70 alone (black trace), together with human CAIV–WT (blue trace), or together with the CAIV mutant H88A. C and D, rate of change in intracellular H+ concentration (Δ[H+]/Δt) during application of lactate (C) and 5% CO2, 10 mm HCO3− (D) in Xenopus oocytes, expressing rMCT2 + rGP70–WT (gray dots), rMCT2 + rGP70–WT + CAIV–WT (blue dots), and rMCT2 + rGP70–WT + CAIV–H88A (red dots), respectively. The black significance indicators above the dots with CAIV refer to the corresponding dots without CAIV (gray dots). The blue significance indicators above the dots for CAIV–H88A refer to the corresponding dots with CAIV–WT (blue dots). E, relative amount of CAIV in CAIV–WT- and CAIV–H88A–expressing oocytes, as determined by Western blot analysis. The inset shows a representative Western blotting against CAIV for Xenopus oocytes expressing CAIV–WT (left lane) and CAIV–H88A (right lane), respectively. F, representative Western blots of CAIV (left blot) and GST (right blot), respectively. CAIV–WT was pulled down with GST (lane 1), and a GST fusion protein of the Ig1 domain of rGP70–WT (lane 2). The mutant CAIV–H88A was pulled down with a GST fusion protein of the Ig1 domain of rGP70–WT (lane 3). Lanes 1 and 2 in the blots are identical to lanes 1 and 2 in Fig. 8D. G, relative intensity of the fluorescent signal of CAIV. For every blot, the signals for CAIV were normalized to the corresponding signals for GST–rGP70 + CAIV–WT. Each individual signal for CAIV was normalized to the intensity of the signal for GST in the same lane. The significance indicators above the dots refer to GST–rGP70 + CAIV–WT.
To test whether the His-88 in CAIV serves as binding site, we pulled down CAIV–WT and CAIV–H88A, with a GST fusion protein of the Ig1 domain of rGP70–WT (Fig. 7F). Although pulldown of CAIV with GST–rGP70–WT again resulted in a signal for CAIV, almost no signal could be observed when GST–rGP70–WT was pulled down with CAIV–H88A (Fig. 7, F and G). This indicates that binding between CAIV and GP70 is indeed mediated by the His-88 in CAIV.
His-88 in CAIV can function both as proton donor and proton acceptor
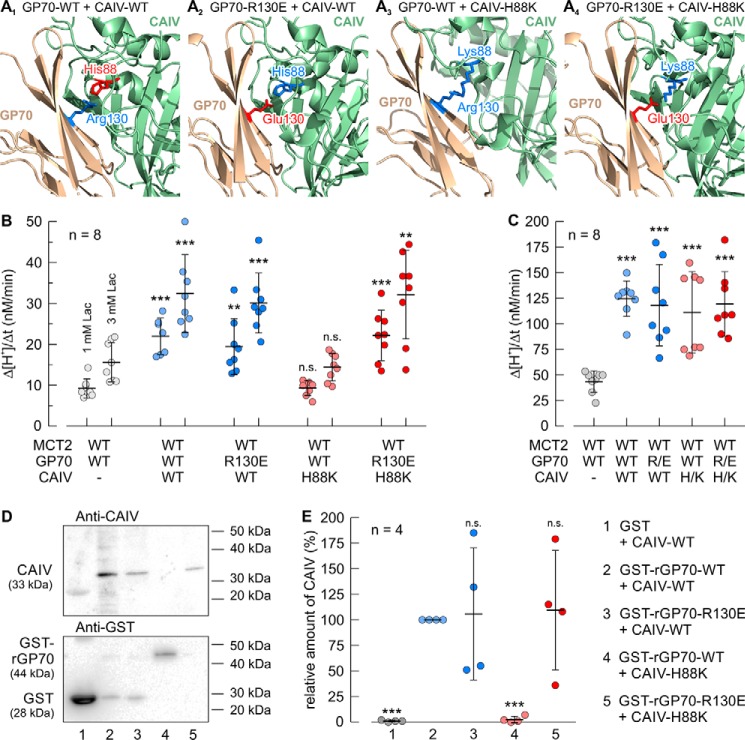
It can be concluded from our results that direct interaction between CAIV and the MCT chaperones CD147 and GP70 is mediated by the His-88 in CAIV and Glu-73, Lys-73, and Arg-130 in hCD147, rCD147, and rGP70, respectively. Although the negatively charged glutamic acid residue could only function as a proton acceptor in a hydrogen bond, the positively charged lysine and arginine residues could only serve as a proton donor. Therefore, His-88 in CAIV has to function both as a proton donor and acceptor, depending on the charge of its binding partner. To test this hypothesis, we combined different mutations in CAIV and rGP70 as depicted in Fig. 8A: Fig. 8A1 shows the interaction between rGP70–WT and CAIV–WT. In this scenario, Arg-130 in rGP70 and the His-88 in CAIV form a hydrogen bond, with the positively charged Arg-130 functioning as proton donor (blue) and His-88 functioning as proton acceptor (red). Fig. 8A2 shows the interaction between rGP70–R130E and CAIV–WT. In this scenario Glu-130 in rGP70 and His-88 in CAIV form a hydrogen bond, with the His-88 now functioning as a proton donor (blue) and the negatively charged Glu-130 functioning as a proton acceptor (red). Fig. 8A3 shows the interaction between rGP70–WT and CAIV–H88K. In this scenario, no hydrogen bond could be formed, because both the Arg-130 in rGP70 and the Lys-88 in CAIV can only function as a proton donor (blue). Fig. 8A4 shows the interaction between rGP70–R130E and CAIV–H88K. In this scenario Glu-130 in rGP70 and Lys-88 in CAIV form a hydrogen bond, with Lys-88 functioning as a proton donor (Fig. 8A4, blue) and Glu-130 functioning as a proton acceptor (red). The four scenarios were first verified experimentally by expression of rMCT2 together with rGP70–WT/R130E and CAIV–WT/H88K in Xenopus oocytes. Transport activity of rMCT2 was determined by measuring Δ[H+]/Δt during application of 1 and 3 mm lactate (Fig. 8B). rMCT2 transport activity was facilitated in all cases where a hydrogen bond could be formed between rGP70 and CAIV (rGP70–WT + CAIV–WT, rGP70–R130E + CAIV–WT, and rGP70–R130E + CAIV–H88K). Only the combination rGP70–WT + CAIV–H88K, where no hydrogen bond could be formed between the two proteins failed to facilitate rMCT2 transport function. Catalytic activity of CAIV was determined by application of 5% CO2, 10 mm HCO3− (Fig. 8C). Coexpression of rMCT2 + rGP70 with both CAIV–WT and CAIV–H88K increased the rate of CO2-induced acidification by a factor of ∼3, with no significant differences between the batches (Fig. 8C). Binding between the different combinations of rGP70 and CAIV was examined by pulldown assay (Fig. 8D). In all cases where a hydrogen bond could be formed between rGP70 and CAIV (rGP70–WT + CAIV–WT, rGP70–R130E + CAIV–WT, and rGP70–R130E + CAIV–H88K), binding between the two proteins could be observed. Only the combination rGP70–WT + CAIV–H88K, where no hydrogen bond could be established between the two proteins, failed to bind (Fig. 8, D and E).
Figure 8.
CAIV–His-88 can function both as a proton donor and a proton acceptor for protein binding. A1–4, structural model of the direct interaction between human CAIV (PDB code 5JN9 (79)) and the Ig1 domain of rat GP70 (based on human CD147, PDB code 3B5H (37)). Amino acids that function as a proton donor are labeled in blue; amino acids that function as proton acceptor are labeled in red. A1, model of the interaction between CAIV–WT and rGP70–WT. In this scenario CAIV–His-88 and rGP70–Arg-130 can form a hydrogen bond with Arg-130 as a proton donor and His-88 as a proton acceptor. A2, model of the interaction between CAIV–WT and rGP70–R130E. In this scenario CAIV–His-88 and rGP70–Glu-130 can form a hydrogen bond with His-88 as proton donor and Glu-130 as proton acceptor. A3, model of the interaction between CAIV–H88K and rGP70–WT. In this scenario, CAIV–Lys-88 and rGP70–Arg-130 cannot form a hydrogen bond because both residues can only function as a proton donor, thereby missing a proton acceptor. A4, model of the interaction between CAIV–H88K and rGP70–R130E. In this scenario CAIV–Lys-88 and rGP70–Glu-130 can again form a hydrogen bond with Lys-88 as a proton donor and Glu-130 as a proton acceptor. B and C, rate of change in intracellular H+ concentration (Δ[H+]/Δt) during application of lactate (B) and CO2/HCO3− (C) in Xenopus oocytes, expressing rMCT2 + rGP70–WT (gray dots), rMCT2 + rGP70–WT + CAIV–WT (light blue dots), rMCT2 + rGP70–R130E + CAIV–WT (dark blue dots), rMCT2 + rGP70–WT + CAIV–H88K (light red dots), and rMCT2 + rGP70–R130E + CAIV–H88K (dark red dots), respectively. The significance indicators above the dots with CAIV refer to the corresponding dots without CAIV (gray dots). D, representative Western blots of CAIV (upper blot) and GST (lower blot), respectively. CAIV–WT was pulled down with GST (lane 1), a GST fusion protein of the Ig1 domain of rGP70–WT (lane 2), and a GST fusion protein of the Ig1 domain of the mutant rGP70–R130E (lane 3). The mutant CAIV–H88K was pulled down with a GST fusion protein of Ig1 domain of rGP70–WT (lane 4), and a GST fusion protein of the Ig1 domain of the mutant rGP70–R130E (lane 5), respectively. Lanes 1 and 2 in the blots are identical to lanes 1 and 2 in Fig. 7F. E, relative intensity of the fluorescent signal of CAIV. For every blot, the signals for CAIV were normalized to the corresponding signals for GST–rGP70 + CAIV–WT. Each individual signal for CAIV was normalized to the intensity of the signal for GST in the same lane. The significance indicators above the dots refer to GST–rGP70–WT + CAIV–WT.
Discussion
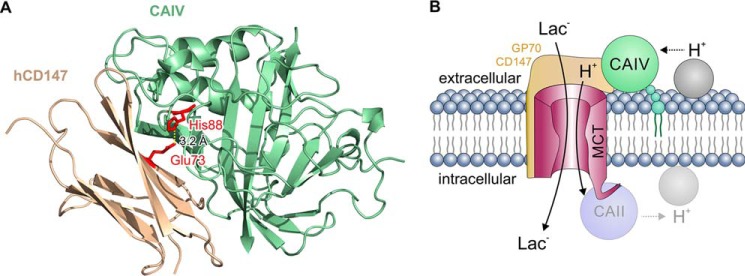
Our previous studies had demonstrated that intracellular CAII binds to MCT1 and MCT4 via a cluster of three glutamic acid residues within the transporter's C-terminal tail (MCT1, 489EEE (62), and MCT4, 431EEE (63)). In both cases, the two outer glutamic acids form hydrogen bonds with the histidine at position 64 in CAII. The extracellular portions of MCT1, MCT2, and MCT4 seem to lack an analogous binding site. Furthermore, CAIV was only able to facilitate transport activity of MCT2 when the transporter was co-expressed with its chaperone GP70 (49). These results suggested that CAIV may interact with MCTs via their chaperones. To investigate whether CAIV directly interacts with the MCT chaperones, we first searched for a putative CAIV-binding site in the human isoform of CD147, the chaperone of MCT1 and MCT4. Molecular docking experiments identified the two glutamic acid residues Glu-31 and Glu-73 in the Ig1 domain of hCD147 as potential interaction partners for CAIV. Subsequent measurements on HEK-293 cells and pulldown experiments revealed that binding of CAIV to hCD147 is mediated by a hydrogen bond between Glu-73 in hCD147 and His-88 in CAIV (Fig. 9A), whereas Glu-31 does not participate in the interaction between the two proteins.
Figure 9.
A, structural model of the direct interaction between CAIV and CD147. CAIV (green cartoon) binds to the Ig1 domain of hCD147 (ochre cartoon) by formation of a hydrogen bond (dotted line) between CAIV–His-88 and CD147–Glu-73 (red sticks) with a distance of 3.2 Å. B, hypothetical model of the functional interaction between MCT, CD147/GP70, and CAIV. CAIV (green circle), which is tethered to the extracellular site of the plasma membrane via a GPI anchor (small green circles), binds MCTs via the Ig1 domain of their chaperones CD147 and GP70 (light ochre structure). This binding brings CAIV close enough to the transporter pore to shuttle protons between transporter and surrounding protonable residues (gray circle). On the intracellular site, CAII (light blue circle), which binds to the C-terminal tail of MCT1 and MCT4 (57, 62, 63), facilitates the exchange of protons between transporter and intracellular protonable residues (light gray circle) in a similar fashion as CAIV. By this noncatalytic mechanism, intracellular and extracellular carbonic anhydrases could facilitate proton-coupled lactate flux across the cell membrane.
Although CAIV–His-88 is highly conserved among all mammalian species, the identity of the amino acid that mediates binding in the chaperone does vary. In fact, binding of human CD147 to CAIV is mediated by the acidic glutamic acid residue at position 73, whereas binding of rat CD147 to CAIV is mediated by an alkaline lysine residue at the same position (rCD147–Lys-73). In rat GP70, binding to CAIV is mediated by the alkaline Arg-130 (which is conserved in the human isoform). These results suggest that hydrogen bond formation with CAIV–His-88 can be mediated by both acidic and alkaline residues. Formation of a hydrogen bond between two residues always requires one residue functioning as a proton donor and one residue functioning as a proton acceptor. Because the negatively charged glutamic acid residue could only function as a proton acceptor in a hydrogen bond, whereas the positively charged lysine and arginine residues could only serve as a proton donor, His-88 in CAIV has to function both as a proton donor and acceptor, depending on the charge of its binding partner. This assumption was confirmed experimentally by systematic exchange of the binding residues in CAIV and rGP70. CAIV–WT could bind to both rGP70–Arg-130 (WT) and rGP70–Glu-130 (mutant). Arg-130 would serve as a proton donor and His-88 as a proton acceptor, whereas Glu-130 would serve as a proton acceptor and His-88 as a proton donor. When His-88 in CAIV was mutated to lysine (which could only function as proton donor), CAIV was only able to bind to rGP70–Glu-130 (proton acceptor) and not to rGP70–Arg-130, because no hydrogen bond could apparently be established between two proton donors.
We cannot fully exclude that introduction of the single-site mutations leads to misfolding of the proteins. However, this scenario seems rather unlikely for several reasons. 1) None of the mutations of the chaperons did influence MCT transport activity in the absence of carbonic anhydrase. If mutation of the Ig1 domain would alter folding of the chaperon, this misfolded chaperon would most likely not be able to interact with MCTs and allow full MCT transport function. 2) As shown in Fig. 8, the effect of single-site mutations in rGP70 can be rescued by mutations in CAIV and vice versa. If the mutation would lead to misfolding of one protein, it would be unlikely that this misfolding could be rescued by mutation of another protein. 3) In case of CAIV, all CAIV mutants show the same catalytic activity as CAIV–WT. It seems unlikely that a misfolded enzyme shows the same catalytic activity as the WT form.
CAIV–H88K was not only able to bind GP70–R130E but also to enhance activity of MCT2 when the transporter was coexpressed with the mutated chaperone. We have previously suggested that CAIV–His-88 (the central residue of the enzyme's intramolecular H+ shuttle) could also mediate H+ transfer between CAIV and MCT (49, 50). Mutation of CAIV–His-88 to lysine should disable H+ shuttling, because lysine has a considerably higher pKa value than the imidazole ring in histidine, which does not allow fast protonation/deprotonation reactions at physiological pH. However, CAIV–H88K does still facilitate MCT2 transport activity (when coexpressed with GP70-R130E) to the same extent as CAIV–WT. This result indicates that His-88 may not be involved in proton transfer between MCT and CAIV, but instead mediates binding of the enzyme to the transporter's chaperone. However, lysine residues that are buried in the protein interior could display considerably lower pKa values than in water (65). Therefore, it is possible that CAIV–H88K retains enough shuttling activity to facilitate MCT-mediated lactate transport. An analogue phenomenon we recently described for the interaction between MCT1/4 and intracellular CAII (57) is as follows. To investigate whether CAII–His-64 mediates binding or proton shuttling between MCT1/4 and CAII, or both, the histidine at position 64 was exchanged either to alanine or to lysine. Mutation of His to Ala should disable both intramolecular H+ shuttling in CAII and binding of the enzyme to the glutamate residues in the C-terminal tail of MCT1/4. Mutation of His to Lys also disables H+ shuttling, but still enables CAII to bind to the transporter. Analogous to these findings, MCT transport activity was not enhanced by CAII–H64A, whereas CAII–H64K facilitated MCT transport activity to the same extent as CAII–WT. These results lead to the conclusion that His-64 may not be involved in proton transfer between MCT and CAII, but instead mediates binding of the enzyme to the transporter. Instead, proton shuttling between transporter and the carbonic anhydrase is mediated by CAII–Glu-69 and CAII–Asp-72, which could function as surface proton antenna for the enzyme (57, 66). For the cancer-associated CAIX, another extracellularly located CA isoform, the proteoglycan-like domain, which is unique to CAIX among the CAs, and which contains a total of 26 glutamate and aspartate residues, was identified as intramolecular H+ shuttle (67). CAIV, however, neither features a proteoglycan-like domain nor has an analogue proton shuttle to CAII–Glu-69/Asp-72 been identified so far. Therefore, it remains unclear which amino acids in CAIV could mediate the proton transfer between MCT and CAIV.
Even though the intermolecular proton shuttle in CAIV has not been identified so far, it appears likely that CAIV facilitates MCT transport activity by an analogous mechanism as described for intracellular CAII and extracellular CAIX. In that case, binding of CAIV to the transporter's chaperone would bring the enzyme close enough to the transporter to rapidly exchange protons between transporter pore and protonable residues at the extracellular face of the plasma membrane. By this mechanism, CAIV would counteract the formation of proton microdomains at the extracellular entrance of the transport pore to facilitate proton-coupled lactate transport across MCTs (Fig. 9B). Indeed, in this study, loss of binding between CAIV and the chaperone always resulted in the loss of CAIV-mediated increase in MCT transport activity. These results show that binding of CAIV to the transporter's chaperone is necessary for the functional interaction between the proteins. However, we cannot fully exclude that the observed increase in MCT transport activity in the presence of CAIV derives from changes the sub-cellular distribution of the transport proteins. Further experiments are required to answer this question.
CAIV also binds to, and enhances, transport activity of several other acid/base transporters. CAIV binds to the 4th extracellular loop of the Cl−/HCO3− anion exchanger AE1 (45). Subsequent mutation studies suggested that the amino acid cluster Ser-643–Leu-655 within this loop has a folded structure that is inaccessible to hydrophilic reagents, whereas the Arg-656–Ile-661 region had an open structure with maximum accessibility at Arg-656 (45). From this it was concluded that CAIV might interact with AE1 within the 656RSEFPI cluster. The exact AE1-binding site in CAIV is yet unknown. However, if we assume that binding between AE1 and CAIV follows the same principle as binding between CD147/GP70 and CAIV, CAIV–His-88 might either bind to Arg-656 or Glu-658 in AE1. Interestingly, both Arg-656 and Glu-658 are subject to point mutations that induce blood group antigens (Moa (R656H), Hga (R656C), and Wra (E658K) (68, 69)). Corresponding to the findings in this study, mutation of AE1–Arg-656 to either cysteine or histidine could be predicted to lead to a loss of binding between AE1 and CAIV, which would result in functional impairment of AE1 transport activity, whereas mutation of AE1–Glu-658 to lysine should still allow binding between transporter and enzyme, without affecting AE1 transport function. Therefore, it appears likely that AE1–Glu-658 serves as binding site for CAIV. These assumptions, of course, require experimental validation.
Pulldown assays demonstrated physical binding between CAIV and a GST fusion protein of the 4th extracellular loop of hNBCe1 (47). However, binding to the loop was abolished when glycine at position 767 was mutated to threonine. Therefore, it was suggested that CAIV binds within the amino acid cluster 766RGW, which is conserved in the bicarbonate transporters (47). According to our present findings, glycine itself would not form a hydrogen bond with the His-88 in CAIV, but the arginine at position 766 might be a potential candidate for the direct interaction with CAIV–His-88. The mutation of Gly-767 to the more bulky threonine might therefore hinder binding of Arg-677 to His-88. This might be interesting to be tested experimentally.
We previously found that CAIV also facilitates transport activity of rMCT1 and rMCT4, when the transporters are expressed without rCD147 in oocytes (50, 51). Xenopus oocytes express endogenous CD147, which has been suggested to interact with the transporters (22). The sequence alignment between the Xenopus CD147 homologue and rat CD147 shows that the Xenopus CD147 features the Thr-75 at the homolog position to rCD147–Lys-73 (Fig. 10). However, threonine should also be able to form a hydrogen bond with CAIV–His-88. Therefore, it is possible that the CAIV-induced facilitation of MCT1/4 transport activity can be mediated by binding of CAIV to the Xenopus CD147 when MCT1/4 are expressed without ancillary protein in the oocytes. It has also been shown that rMCT2 can interact with endogenous neuroplastin, when heterologously expressed in Xenopus oocytes (70). The Xenopus neuroplastin homologue, which has 39% sequence identity with rCD147, features Glu-94 at the position analogue to Lys-73 in rCD147 (Fig. 10). However, we have previously shown that transport activity of MCT2 can only be enhanced by CAIV when the transporter was coexpressed with rGP70 in oocytes, but not when MCT2 and CAIV were coexpressed without the ancillary protein (49). It appears questionable that CAIV can directly interact with endogenous neuroplastin to facilitate MCT2 transport activity in Xenopus oocytes.
Figure 10.

Alignment of the amino acid sequence surrounding the putative CAIV-binding site of rat CD147 (AAH59145.1), Xenopus CD147 orthologue (NP_001089604.1), and the Xenopus neuroplastin orthologue (NP_001082482). The putative CAIV-binding site is marked in red.
Experimental procedures
Molecular docking of CD147/GP70 and CAIV
There is no solved crystal structure of either rCD147 or rGP70, and therefore, in silico models were generated based on their respective sequences. The on-line software Swiss-Model was used to generate the rCD147 and rGP70 Ig domain models utilizing hCD147 (PDB code 3B5H) as the template. Swiss-Model generates protein models based on previously solved structures of highly homologous proteins (71). Using the interactive graphical program Coot (72), hCD147 (PDB code 3B5H) and rGP70 models were manually docked onto to CAIV (PDB code 5JN9) active-site domain to maximize surface interactions. Energy minimization was then performed in the crystallography and NMR system program (CNS) to achieve a local minimum energy at the complex interface (73). Complexes were further analyzed, and figures were generated using the graphics software PyMOL (Schrödinger).
Constructs
Human CD147, cloned into the mammalian vector pCMV6-Entry, was purchased from OriGene Technologies (RC203894). The construct carries a C-terminal Myc-DDK tag. Rat CD147 and rat GP70, both cloned into oocyte expression vector pGEM-He-Juel, which contains the 5′- and the 3′-untranscribed regions of the Xenopus β-globin, flanking the multiple cloning site, were kindly provided by Dr. Andrew Halestrap, University of Bristol, UK (21, 25). Rat MCT1 and rat MCT2, cloned into oocyte expression vector pGEM-He-Juel, were provided by Dr. Stefan Bröer, Australian National University, Canberra (2, 8). Human CAIV was provided by Dr. William S. Sly, St. Louis, MO, and subcloned into pGEM-He-Juel, as described previously (49).
Site-directed mutation
Site-directed mutation of hCD147, rCD147, rGP70, and CAIV was carried out by PCR using Phusion High-Fidelity DNA polymerase (ThermoFisher Scientific) and modified primers, which contained the desired mutation. Primers used for creation of the different mutants are shown in Table 1. hCD147, cloned into the expression vector pCMV6-Entry, rCD147, rGP70, and CAIV, cloned into the oocyte expression vector pGEM-He-Juel, were used as template. PCR was cleaned up using the GeneJET PCR purification kit (ThermoFisher Scientific), and the template was digested with DpnI (Fermentas FastDigest DpnI, ThermoFisher Scientific) before transformation into Escherichia coli DH5α cells. Successful mutation of the construct was confirmed by sequencing (SEQ-IT; Macrogen Europe).
Table 1.
Primers used to generate mutants
Shown are the sense primers for single-site mutation of hCD147, rCD147, rGP70, and CAIV. Nucleotides that differ from the wildtype sequence are labeled in bold. The antisense primers had the inverse complement sequence of the sense primers.
| Mutant | Primer sequence (5′ → 3′) |
|---|---|
| hCD147–E31Q | CAGTCTTCACTACCGTACAAGACCTTGGCTCCAAG |
| hCD147–E73Q | CCGGCCAGAAAACGCAGTTCAAGGTGGACTC |
| rCD147–E32A | GTAACCTCTGTCCAGGCAGTTGACTCCAAGACACAG |
| rCD147–K73A | CCCGATCTACAGATGGCGTACACGGTGGATGC |
| rGP70-D95A | CGTTCACGGCAACTGAGGCTGTGATGTCAATG |
| rGP70–R130A | GGGGACACCTTATACAGTCAATACGCGTTCACCG |
| rGP70–R130E | GGACACCTTATACAGTCAATACGAATTCACCG |
| CAIV–H88A | GGACTGTCCAAAATAACGGGGCCTCAGTGATG |
| CAIV–H88K | GGACTGTCCAAAATAACGGGAAGTCAGTGATG |
Protein expression in HEK-293 cells
2.8 μg of cDNA encoding hCD147–WT, hCD147–E31Q, hCD147–E73Q, and hCD147–E31Q/E73Q (cloned into pCMV6-Entry) and CAIV (cloned into pcDNA3) were transiently transfected using the calcium phosphate method (74). HEK-293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% (v/v) fetal bovine serum, 5% (v/v) calf serum, and 1% (v/v) penicillin/streptomycin/glutamine. Cells were incubated at 37 °C in 5% CO2, 95% air in humidified cell culture incubators. Experiments were carried out 42–56 h after transfection. At this time, cells reached maximum DNA expression and 85–90% confluency.
Measurement of intracellular H+ concentration in HEK-293 cells
Transport activity of MCT1 was measured by a fluorescent assay, slightly modified from Ref. 75. In brief, HEK-293 cells, grown on poly-l-lysine–coated coverslips (placed on 100-mm dishes), were transiently transfected with cDNA as described above. Two days after transfection, coverslips were rinsed in serum-free DMEM and incubated in 2 ml of serum-free DMEM, containing 2 μm 2′,7′-bis(2-carboxyethyl)-5-(and -6)-carboxyfluorescein, acetoxymethyl ester (BCECF-AM) at room temperature for 15 min in the dark. Coverslips were then mounted in a fluorescence cuvette and were perfused with HEPES-buffered solution (140 mm NaCl, 25 mm sodium gluconate, 5 mm potassium gluconate, 1 mm calcium gluconate, 5 mm glucose, 1 mm MgSO4, 10 mm HEPES, 2.5 mm NaH2PO4, pH 7.4) in the nominal absence of CO2/HCO3−. In lactate-containing saline, NaCl was replaced by an equimolar amount of sodium l-lactate. To check the expression of CAIV, a short CO2 pulse was applied. For this solution, sodium gluconate was replaced by 25 mm NaHCO3, and the solution was bubbled with air containing 5% CO2.
Fluorescence was monitored using an RCR fluorometer (Photon Technologies International Inc.) at excitation wavelengths of 440 and 502.5 nm and an emission wavelength of 528.7 nm. The system was calibrated by the use of nigericin (1 μm). The rate of change in [H+]i was calculated by determining the slope at each lactate or CO2/HCO3− application. All calculations were performed with the program OriginPro 8.6 (OriginLab Corp.).
Analysis of protein expression in HEK-293 cells by Western blotting
Two days after transfection, cells were lysed in immunoprecipitation buffer (10 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 5 mm EDTA, 0.15 mm NaCl, 0.5% sodium deoxycholate), containing complete protease inhibitor mixture and 5 mm phenylmethylsulfonyl fluoride. The protein concentration was determined by BCA assay. 50 μg of each sample were loaded per lane on a 12% acrylamide gel and transferred to nitrocellulose membranes. Proteins of interest were labeled with rabbit anti-MCT1 (1:200, AB-3538p, Millipore), mouse anti-MYC (1:4000, 05-724, Millipore), mouse anti-CD147 (1:500, sc-21746 Santa Cruz Biotechnology), and mouse anti-GAPDH (1:5000, SC-47724, Santa Cruz Biotechnology), followed by incubation with horseradish peroxidase-conjugated secondary antibodies (1:1000, goat anti-rabbit and sheep anti-mouse, Santa Cruz Biotechnology). Immunoblots were developed with Luminata Crescendo Western horseradish peroxidase reagent (Millipore) and visualized using a biomolecular imager (ImageQuant LAS4000, GE Healthcare). Quantitative densitometry analyses were performed with ImageJ. To allow comparison of different Western blottings, all measured protein concentrations on one blot were normalized to the concentration of GAPDH on the same blot.
Heterologous protein expression in Xenopus oocytes
cDNA coding for human CAIV, rat MCT1, rat MCT2, rat CD147, and rat GP70, respectively (all cloned into the oocyte expression vector pGEM-He-Juel), was transcribed in vitro with T7 RNA-polymerase (mMessage mMachine, Ambion Inc., Austin, TX), as described earlier (76, 77). Xenopus laevis females were purchased from the Radboud University, Nijmegen, Netherlands. Segments of ovarian lobules were surgically removed under sterile conditions from frogs anesthetized with 1 g/liter ethyl 3-aminobenzoate methanesulfonate (MS-222, Sigma) and rendered hypothermic. The procedure was approved by the Landesuntersuchungsamt Rheinland-Pfalz, Koblenz (23 177-07/A07-2-003 §6) and the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg (33.19-42502-05-17A113). As described earlier (76, 77), oocytes were singularized by collagenase (Collagenase A, Roche Applied Science) treatment in Ca2+-free oocyte saline (pH 7.8) at 28 °C for 2 h. The singularized oocytes were left overnight in an incubator at 18 °C in Ca2+-containing oocyte saline (pH 7.8) to recover. Oocytes of stages V and VI were injected with 5 ng of cRNA coding for rMCT1 or rMCT2, together with 10 ng of cRNA coding for rCD147–WT or a mutant of rCD147, or 10 ng of cRNA coding for rGP70–WT or a mutant of rGP70, and 1 ng of cRNA coding for CAIV–WT or a mutant of CAIV. Measurements were carried out 3–6 days after injection of cRNA. The oocyte saline had the following composition: 82.5 mm NaCl, 2.5 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 1 mm Na2HPO4, 5 mm HEPES, titrated with NaOH to the desired pH.
Measurement of intracellular H+ concentration in Xenopus oocytes
All measurements were carried out in oocyte saline (pH 7.0) in the nominal absence of CO2/HCO3−, containing around 0.008 mm CO2 from air and hence a HCO3− concentration of less than 0.2 mm. In lactate-containing saline, NaCl was replaced by an equivalent amount of sodium l-lactate. To check for CAIV catalytic activity, a short CO2 pulse was applied. For this solution, NaCl was replaced by 10 mm NaHCO3, and the solution was aerated with 5% CO2, 95% O2.
To measure intracellular H+ concentration and membrane potential, single-barreled microelectrodes were used; the manufacture and application have been described in detail previously (77, 78). Briefly, a borosilicate glass capillary of 1.5 mm in diameter was pulled to a micropipette and was silanized with a drop of 5% tri-N-butylchlorosilane in 99.9% pure carbon tetrachloride, backfilled into the tip. The micropipette was baked for 4.5 min at 450 °C on a hot plate. H+-sensitive mixture (Fluka 95291, Fluka) was backfilled into the silanized tip and filled up with 0.1 m sodium citrate, pH 6.0. To increase the opening of the electrode tip, it was beveled with a jet stream of aluminum powder suspended in H2O. The reference electrode was filled with 3 m KCl. Calibration of the electrodes was carried out in oocyte salines with a pH of 7.0 and 6.4. As described previously (2), optimal pH changes were detected when the electrode was located near the inner surface of the plasma membrane. During all measurements, oocytes were clamped to a holding potential of −40 mV using an additional microelectrode, filled with 3 m KCl and connected to an Axoclamp 2B amplifier (Axon Instruments). All experiments were carried out at room temperature (22–25 °C). The measurements were stored digitally using custom-made PC software based on the program LabView (National Instruments). The rate of change of the measured [H+]i was analyzed by determining the slope of a linear regression fit using the spreadsheet program OriginPro 8.6 (OriginLab Corporation). Conversion and analysis of the data have been described in detail previously (77).
Pulldown of CAIV with GST fusion proteins
To create GST fusion proteins, cDNA sequences encoding the Ig1 domain of hCD147–WT (amino acids 23–105), rCD147–WT (amino acids 23–105), rGP70–WT (amino acids 24–163), or mutants of the chaperones were subcloned into the bacterial expression vector pGEX-2T (GE Healthcare) and transformed into E. coli BL21 cells. Protein expression was induced by addition of 0.8 mm isopropyl β-d-thiogalactopyranoside. 3 h after induction, cells were harvested, resuspended in PBS, supplemented with 2 mm MgCl2 and protease inhibitor mixture tablets (PBS+), and lysed by addition of 1 volume of lysis buffer (catalog no. 21516, ThermoFisher Scientific). Bacterial lysates were centrifuged for 15 min at 4 °C, 12,000 × g, and the supernatant, containing the GST fusion protein (bait protein), was collected for further use. CAIV–WT and mutants of CAIV, respectively, were expressed in Xenopus oocytes. For each experiment, 20 oocytes were lysed in PBS+ + lysis buffer. Oocyte lysates were centrifuged for 15 min at 4 °C, 12,000 × g, and the supernatant (prey protein) was collected for further use.
The pulldown experiment was carried out using the Pierce GST protein interaction pulldown kit (ThermoFisher Scientific). Briefly, for immobilization of GST fusion protein, bacterial lysate was added to GSH-agarose and incubated for 2 h at 4 °C with end-over-end mixing. After incubation, the excess bait protein was removed by centrifugation, and the beads were washed five times with PBS+. 400 μl of oocyte lysate, containing CAIV, was added to the column and incubated overnight at 4 °C with end-over-end mixing. After incubation, the excess prey protein was removed by centrifugation, and the beads were washed two times with wash buffer. Protein was eluted from the beads with 250 μl of elution buffer (10 mm GSH in PBS, pH 8.0).
To determine the relative amount of GST and CAIV, an equal volume of the samples was analyzed by Western blotting. GST was detected using a primary anti-GST antibody (dilution 1:400; anti-GST tag mouse monoclonal IgG, 05-782, Millipore) and a goat anti-mouse IgG horseradish peroxidase-conjugated secondary antibody (dilution 1:2000; sc-2031, Santa Cruz Biotechnology). CAIV was detected using primary anti-CAIV antibody (1:300 mouse anti-human CAIV mAb, MAB2186, R&D Systems) and a goat anti-mouse IgG horseradish peroxidase-conjugated secondary antibody (dilution 1:2000; Santa Cruz Biotechnology).
Imaging of the blots was carried out with an Odyssey® Fc Imaging system (LI-COR Inc.) and the software Image StudioTM. Optimal exposure times were automatically selected by the software, and all bands were controlled for overexposure. Quantification of the band intensity was carried out with the software ImageJ. To overcome variations in the signal intensity between different blots, signal intensity of each band for CAIV was normalized to the signal intensity of the band from the pulldown of CAIV–WT with the GST fusion protein of the WT chaperone on the same blot. To account for variations in the amount of GST fusion protein, each normalized signal for CAIV was divided by the corresponding normalized signal for GST.
Calculation and statistics
Statistical values are presented as means ± S.D. For calculation of significance in differences, Student's t test was used. In the figures shown, a significance level of p ≤ 0.05 is marked with *, p ≤ 0.01 is marked with **, and p ≤ 0.001 is marked with ***.
Author contributions
L. S. F.-Q., R. M., J. R. C., J. W. D., and H. M. B. conceptualization; L. S. F.-Q., S. A., H.-P. S., A. T., C. D. B., J. T. A., and H. M. B. investigation; L. S. F.-Q., S. A., J. T. A., R. M., J. R. C., and J. W. D. writing-review and editing; R. M., J. R. C., J. W. D., and H. M. B. supervision; J. R. C., J. W. D., and H. M. B. funding acquisition; J. W. D. and H. M. B. project administration; H. M. B. visualization; H. M. B. writing-original draft.
This work was supported by Deutsche Forschungsgemeinschaft Grant BE 4310/6-1 (to H. M. B.), International Research Training Group 1830 (to L. S. F.-Q., J. R. C., J. W. D., and H. M. B.), Stiftung Rheinland-Pfalz für Innovation Grant 961-386261/957 (to H. M. B.), the Research Initiative BioComp (to J. W. D. and H. M. B.), and the Landesschwerpunkt Membrantransport (to J. W. D. and H. M. B.). The authors declare that they have no conflicts of interest with the contents of this article.
- MCT
- monocarboxylate transporter
- CAIV
- carbonic anhydrase isoform IV
- CD147
- cluster of differentiation 147 (also termed basigin or EMMPRIN)
- GP70
- glycoprotein 70 (also termed embigin)
- Ig domain
- immunoglobulin-like domain
- DMEM
- Dulbecco's modified Eagle's medium
- PDB
- Protein Data Bank
- CA
- carbonic anhydrase
- GPI
- glycosylphosphatidylinositol
- GST
- glutathione S-transferase.
References
- 1. Bröer S., Rahman B., Pellegri G., Pellerin L., Martin J. L., Verleysdonk S., Hamprecht B., and Magistretti P. J. (1997) Comparison of lactate transport in astroglial cells and monocarboxylate transporter 1 (MCT 1) expressing Xenopus laevis oocytes. Expression of two different monocarboxylate transporters in astroglial cells and neurons. J. Biol. Chem. 272, 30096–30102 10.1074/jbc.272.48.30096 [DOI] [PubMed] [Google Scholar]
- 2. Bröer S., Schneider H. P., Bröer A., Rahman B., Hamprecht B., and Deitmer J. W. (1998) Characterization of the monocarboxylate transporter 1 expressed in Xenopus laevis oocytes by changes in cytosolic pH. Biochem. J. 333, 167–174 10.1042/bj3330167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Halestrap A. P., and Price N. T. (1999) The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem. J. 343, 281–299 10.1042/0264-6021:3430281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Halestrap A. P. (2013) The SLC16 gene family-structure, role and regulation in health and disease. Mol. Aspects Med. 34, 337–349 10.1016/j.mam.2012.05.003 [DOI] [PubMed] [Google Scholar]
- 5. Garcia C. K., Brown M. S., Pathak R. K., and Goldstein J. L. (1995) cDNA cloning of MCT2, a second monocarboxylate transporter expressed in different cells than MCT1. J. Biol. Chem. 270, 1843–1849 10.1074/jbc.270.4.1843 [DOI] [PubMed] [Google Scholar]
- 6. Jackson V. N., and Halestrap A. P. (1996) The kinetics, substrate, and inhibitor specificity of the monocarboxylate (lactate) transporter of rat liver cells determined using the fluorescent intracellular pH indicator, 2′,7′-bis(carboxyethyl)-5(6)-carboxyfluorescein. J. Biol. Chem. 271, 861–868 10.1074/jbc.271.2.861 [DOI] [PubMed] [Google Scholar]
- 7. Jackson V. N., Price N. T., Carpenter L., and Halestrap A. P. (1997) Cloning of the monocarboxylate transporter isoform MCT2 from rat testis provides evidence that expression in tissues is species-specific and may involve post-transcriptional regulation. Biochem. J. 324, 447–453 10.1042/bj3240447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bröer S., Bröer A., Schneider H. P., Stegen C., Halestrap A. P., and Deitmer J. W. (1999) Characterization of the high-affinity monocarboxylate transporter MCT2 in Xenopus laevis oocytes. Biochem. J. 341, 529–535 10.1042/bj3410529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pellerin L., Pellegri G., Bittar P. G., Charnay Y., Bouras C., Martin J. L., Stella N., and Magistretti P. J. (1998) Evidence supporting the existence of an activity-dependent astrocyte-neuron lactate shuttle. Dev. Neurosci. 20, 291–299 10.1159/000017324 [DOI] [PubMed] [Google Scholar]
- 10. Pellerin L., Pellegri G., Martin J.-L., and Magistretti P. J. (1998) Expression of monocarboxylate transporter mRNAs in mouse brain: support for a distinct role of lactate as an energy substrate for the neonatal vs. adult brain. Proc. Natl. Acad. Sci. U.S.A. 95, 3990–3995 10.1073/pnas.95.7.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergersen L., Waerhaug O., Helm J., Thomas M., Laake P., Davies A. J., Wilson M. C., Halestrap A. P., and Ottersen O. P. (2001) A novel postsynaptic density protein: the monocarboxylate transporter MCT2 is co-localized with δ-glutamate receptors in postsynaptic densities of parallel fiber-Purkinje cell synapses. Exp. Brain Res. 136, 523–534 10.1007/s002210000600 [DOI] [PubMed] [Google Scholar]
- 12. Pierre K., and Pellerin L. (2005) Monocarboxylate transporters in the central nervous system: distribution, regulation and function. J. Neurochem. 94, 1–14 10.1111/j.1471-4159.2005.03168.x [DOI] [PubMed] [Google Scholar]
- 13. Yoon H., Fanelli A., Grollman E. F., and Philp N. J. (1997) Identification of a unique monocarboxylate transporter (MCT3) in retinal pigment epithelium. Biochem. Biophys. Res. Commun. 234, 90–94 10.1006/bbrc.1997.6588 [DOI] [PubMed] [Google Scholar]
- 14. Philp N. J., Yoon H., and Grollman E. F. (1998) Monocarboxylate transporter MCT1 is located in the apical membrane and MCT3 in the basal membrane of rat RPE. Am. J. Physiol. 274, R1824–R1828 [DOI] [PubMed] [Google Scholar]
- 15. Philp N. J., Yoon H., and Lombardi L. (2001) Mouse MCT3 gene is expressed preferentially in retinal pigment and choroid plexus epithelia. Am. J. Physiol. Cell Physiol. 280, C1319–C1326 10.1152/ajpcell.2001.280.5.C1319 [DOI] [PubMed] [Google Scholar]
- 16. Grollman E. F., Philp N. J., McPhie P., Ward R. D., and Sauer B. (2000) Determination of transport kinetics of chick MCT3 monocarboxylate transporter from retinal pigment epithelium by expression in genetically modified yeast. Biochemistry 39, 9351–9357 10.1021/bi000464+ [DOI] [PubMed] [Google Scholar]
- 17. Dimmer K. S., Friedrich B., Lang F., Deitmer J. W., and Bröer S. (2000) The low-affinity monocarboxylate transporter MCT4 is adapted to the export of lactate in highly glycolytic cells. Biochem. J. 350, 219–227 10.1042/bj3500219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pinheiro C., Longatto-Filho A., Azevedo-Silva J., Casal M., Schmitt F. C., and Baltazar F. (2012) Role of monocarboxylate transporters in human cancers: state of the art. J. Bioenerg. Biomembr. 44, 127–139 10.1007/s10863-012-9428-1 [DOI] [PubMed] [Google Scholar]
- 19. Pinheiro C., Reis R. M., Ricardo S., Longatto-Filho A., Schmitt F., and Baltazar F. (2010) Expression of monocarboxylate transporters 1, 2, and 4 in human tumours and their association with CD147 and CD44. J. Biomed. Biotechnol. 2010, 427694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kirk P., Wilson M. C., Heddle C., Brown M. H., Barclay A. N., and Halestrap A. P. (2000) CD147 is tightly associated with lactate transporters MCT1 and MCT4 and facilitates their cell surface expression. EMBO J. 19, 3896–3904 10.1093/emboj/19.15.3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilson M. C., Meredith D., and Halestrap A. P. (2002) Fluorescence resonance energy transfer studies on the interaction between the lactate transporter MCT1 and CD147 provide information on the topology and stoichiometry of the complex in situ. J. Biol. Chem. 277, 3666–3672 10.1074/jbc.M109658200 [DOI] [PubMed] [Google Scholar]
- 22. Wilson M. C., Meredith D., Fox J. E., Manoharan C., Davies A. J., and Halestrap A. P. (2005) Basigin (CD147) is the target for organomercurial inhibition of monocarboxylate transporter isoforms 1 and 4: the ancillary protein for the insensitive MCT2 is embigin (gp70). J. Biol. Chem. 280, 27213–27221 10.1074/jbc.M411950200 [DOI] [PubMed] [Google Scholar]
- 23. Halestrap A. P. (2012) The monocarboxylate transporter family-structure and functional characterization. IUBMB Life 64, 1–9 10.1002/iub.573 [DOI] [PubMed] [Google Scholar]
- 24. Manoharan C., Wilson M. C., Sessions R. B., and Halestrap A. P. (2006) The role of charged residues in the transmembrane helices of monocarboxylate transporter 1 and its ancillary protein basigin in determining plasma membrane expression and catalytic activity. Mol. Membr. Biol. 23, 486–498 10.1080/09687860600841967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ovens M. J., Manoharan C., Wilson M. C., Murray C. M., and Halestrap A. P. (2010) The inhibition of monocarboxylate transporter 2 (MCT2) by AR-C155858 is modulated by the associated ancillary protein. Biochem. J. 431, 217–225 10.1042/BJ20100890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilson M. C., Meredith D., Bunnun C., Sessions R. B., and Halestrap A. P. (2009) Studies on the DIDS-binding site of monocarboxylate transporter 1 suggest a homology model of the open conformation and a plausible translocation cycle. J. Biol. Chem. 284, 20011–20021 10.1074/jbc.M109.014217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Igakura T., Kadomatsu K., Kaname T., Muramatsu H., Fan Q. W., Miyauchi T., Toyama Y., Kuno N., Yuasa S., Takahashi M., Senda T., Taguchi O., Yamamura K., Arimura K., and Muramatsu T. (1998) A null mutation in basigin, an immunoglobulin superfamily member, indicates its important roles in peri-implantation development and spermatogenesis. Dev. Biol. 194, 152–165 10.1006/dbio.1997.8819 [DOI] [PubMed] [Google Scholar]
- 28. Saxena D. K., Oh-Oka T., Kadomatsu K., Muramatsu T., and Toshimori K. (2002) Behaviour of a sperm surface transmembrane glycoprotein basigin during epididymal maturation and its role in fertilization in mice. Reproduction 123, 435–444 10.1530/rep.0.1230435 [DOI] [PubMed] [Google Scholar]
- 29. Schlosshauer B. (1991) Neurothelin: molecular characteristics and developmental regulation in the chick CNS. Development 113, 129–140 [DOI] [PubMed] [Google Scholar]
- 30. Fadool J. M., and Linser P. J. (1993) 5A11 antigen is a cell recognition molecule which is involved in neuronal–glial interactions in avian neural retina. Dev. Dyn. 196, 252–262 10.1002/aja.1001960406 [DOI] [PubMed] [Google Scholar]
- 31. Pushkarsky T., Yurchenko V., Laborico A., and Bukrinsky M. (2007) CD147 stimulates HIV-1 infection in a signal-independent fashion. Biochem. Biophys. Res. Commun. 363, 495–499 10.1016/j.bbrc.2007.08.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muramatsu T., and Miyauchi T. (2003) Basigin (CD147): a multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion. Histol. Histopathol. 18, 981–987 [DOI] [PubMed] [Google Scholar]
- 33. Nabeshima K., Iwasaki H., Koga K., Hojo H., Suzumiya J., and Kikuchi M. (2006) Emmprin (basigin/CD147): matrix metalloproteinase modulator and multifunctional cell recognition molecule that plays a critical role in cancer progression. Pathol. Int. 56, 359–367 10.1111/j.1440-1827.2006.01972.x [DOI] [PubMed] [Google Scholar]
- 34. Fossum S., Mallett S., and Barclay A. N. (1991) The MRC OX-47 antigen is a member of the immunoglobulin superfamily with an unusual transmembrane sequence. Eur. J. Immunol. 21, 671–679 10.1002/eji.1830210320 [DOI] [PubMed] [Google Scholar]
- 35. Redzic J. S., Armstrong G. S., Isern N. G., Jones D. N., Kieft J. S., and Eisenmesser E. Z. (2011) The retinal specific CD147 Ig0 domain: From molecular structure to biological activity. J. Mol. Biol. 411, 68–82 10.1016/j.jmb.2011.04.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Muramatsu T. (2016) Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J. Biochem. 159, 481–490 10.1093/jb/mvv127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu X. L., Hu T., Du J. M., Ding J. P., Yang X. M., Zhang J., Yang B., Shen X., Zhang Z., Zhong W. D., Wen N., Jiang H., Zhu P., and Chen Z. N. (2008) Crystal structure of HAb18G/CD147: implications for immunoglobulin superfamily homophilic adhesion. J. Biol. Chem. 283, 18056–18065 10.1074/jbc.M802694200 [DOI] [PubMed] [Google Scholar]
- 38. Guenette R. S., Sridhar S., Herley M., Mooibroek M., Wong P., and Tenniswood M. (1997) Embigin, A developmentally expressed member of the immunoglobulin super family, is also expressed during regression of prostate and mammary gland. Dev. Genet. 21, 268–278 10.1002/(SICI)1520-6408(1997)21:4%3C268::AID-DVG4%3E3.0.CO%3B2-5 [DOI] [PubMed] [Google Scholar]
- 39. Lain E., Carnejac S., Escher P., Wilson M. C., Lømo T., Gajendran N., and Brenner H. R. (2009) A novel role for embigin to promote sprouting of motor nerve terminals at the neuromuscular junction. J. Biol. Chem. 284, 8930–8939 10.1074/jbc.M809491200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jung D. E., Kim J. M., Kim C., and Song S. Y. (2016) Embigin is overexpressed in pancreatic ductal adenocarcinoma and regulates cell motility through epithelial to mesenchymal transition via the TGF-β pathway. Mol. Carcinog. 55, 633–645 10.1002/mc.22309 [DOI] [PubMed] [Google Scholar]
- 41. Chao F., Zhang J., Zhang Y., Liu H., Yang C., Wang J., Guo Y., Wen X., Zhang K., Huang B., Liu D., and Li Y. (2015) Embigin, regulated by HOXC8, plays a suppressive role in breast tumorigenesis. Oncotarget 6, 23496–23509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maren T. H. (1967) Carbonic anhydrase: chemistry, physiology, and inhibition. Physiol. Rev. 47, 595–781 10.1152/physrev.1967.47.4.595 [DOI] [PubMed] [Google Scholar]
- 43. Zhu X. L., and Sly W. S. (1990) Carbonic anhydrase IV from human lung. Purification, characterization, and comparison with membrane carbonic anhydrase from human kidney. J. Biol. Chem. 265, 8795–8801 [PubMed] [Google Scholar]
- 44. Sterling D., and Casey J. R. (2002) Bicarbonate transport proteins. Biochem. Cell Biol. 80, 483–497 10.1139/o02-152 [DOI] [PubMed] [Google Scholar]
- 45. Sterling D., Alvarez B. V., and Casey J. R. (2002) The extracellular component of a transport metabolon: extracellular loop 4 of the human AE1 Cl−/HCO3− exchanger binds carbonic anhydrase IV. J. Biol. Chem. 277, 25239–25246 10.1074/jbc.M202562200 [DOI] [PubMed] [Google Scholar]
- 46. Svichar N., Waheed A., Sly W. S., Hennings J. C., Hübner C. A., and Chesler M. (2009) Carbonic anhydrases CA4 and CA14 both enhance AE3-mediated Cl−-HCO3− exchange in hippocampal neurons. J. Neurosci. 29, 3252–3258 10.1523/JNEUROSCI.0036-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alvarez B. V., Loiselle F. B., Supuran C. T., Schwartz G. J., and Casey J. R. (2003) Direct extracellular interaction between carbonic anhydrase IV and the human NBC1 sodium/bicarbonate co-transporter. Biochemistry 42, 12321–12329 10.1021/bi0353124 [DOI] [PubMed] [Google Scholar]
- 48. Wu Q., Pierce W. M. Jr., and Delamere N. A. (1998) Cytoplasmic pH responses to carbonic anhydrase inhibitors in cultured rabbit nonpigmented ciliary epithelium. J. Membr. Biol. 162, 31–38 10.1007/s002329900339 [DOI] [PubMed] [Google Scholar]
- 49. Klier M., Schüler C., Halestrap A. P., Sly W. S., Deitmer J. W., and Becker H. M. (2011) Transport activity of the high-affinity monocarboxylate transporter MCT2 is enhanced by extracellular carbonic anhydrase IV but not by intracellular carbonic anhydrase II. J. Biol. Chem. 286, 27781–27791 10.1074/jbc.M111.255331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Klier M., Andes F. T., Deitmer J. W., and Becker H. M. (2014) Intracellular and extracellular carbonic anhydrases cooperate non-enzymatically to enhance activity of monocarboxylate transporters. J. Biol. Chem. 289, 2765–2775 10.1074/jbc.M113.537043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Noor S. I., Pouyssegur J., Deitmer J. W., and Becker H. M. (2017) Integration of a 'proton antenna' facilitates transport activity of the monocarboxylate transporter MCT4. FEBS J. 284, 149–162 10.1111/febs.13964 [DOI] [PubMed] [Google Scholar]
- 52. Becker H. M., Hirnet D., Fecher-Trost C., Sültemeyer D., and Deitmer J. W. (2005) Transport activity of MCT1 expressed in Xenopus oocytes is increased by interaction with carbonic anhydrase. J. Biol. Chem. 280, 39882–39889 10.1074/jbc.M503081200 [DOI] [PubMed] [Google Scholar]
- 53. Becker H. M., and Deitmer J. W. (2008) Nonenzymatic proton handling by carbonic anhydrase II during H+-lactate cotransport via monocarboxylate transporter 1. J. Biol. Chem. 283, 21655–21667 10.1074/jbc.M802134200 [DOI] [PubMed] [Google Scholar]
- 54. Becker H. M., Klier M., and Deitmer J. W. (2010) Nonenzymatic augmentation of lactate transport via monocarboxylate transporter isoform 4 by carbonic anhydrase II. J. Membr. Biol. 234, 125–135 10.1007/s00232-010-9240-y [DOI] [PubMed] [Google Scholar]
- 55. Becker H. M., Klier M., Schüler C., McKenna R., and Deitmer J. W. (2011) Intramolecular proton shuttle supports not only catalytic but also noncatalytic function of carbonic anhydrase II. Proc. Natl. Acad. Sci. U.S.A. 108, 3071–3076 10.1073/pnas.1014293108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Almquist J., Lang P., Prätzel-Wolters D., Deitmer J. W., Jirstrand M., and Becker H. M. (2010) A kinetic model of the monocarboxylate transporter MCT1 and its interaction with carbonic anhydrase II. J. Comput. Sci. Syst. Biol. 3, 107–116 [Google Scholar]
- 57. Noor S. I., Jamali S., Ames S., Langer S., Deitmer J. W., and Becker H. M. (2018) A surface proton antenna in carbonic anhydrase II supports lactate transport in cancer cells. Elife 7, e35176 10.7554/eLife.35176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Adelroth P., and Brzezinski P. (2004) Surface-mediated proton-transfer reactions in membrane-bound proteins. Biochim. Biophys. Acta 1655, 102–115 10.1016/j.bbabio.2003.10.018 [DOI] [PubMed] [Google Scholar]
- 59. Friedman R., Nachliel E., and Gutman M. (2005) Molecular dynamics of a protein surface: ion-residues interactions. Biophys. J. 89, 768–781 10.1529/biophysj.105.058917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brändén M., Sandén T., Brzezinski P., and Widengren J. (2006) Localized proton microcircuits at the biological membrane-water interface. Proc. Natl. Acad. Sci. U.S.A. 103, 19766–19770 10.1073/pnas.0605909103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martínez C., Kalise D., and Barros L. F. (2010) General requirement for harvesting antennae at Ca and H channels and transporters. Front. Neuroenergetics 2, 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stridh M. H., Alt M. D., Wittmann S., Heidtmann H., Aggarwal M., Riederer B., Seidler U., Wennemuth G., McKenna R., Deitmer J. W., and Becker H. M. (2012) Lactate flux in astrocytes is enhanced by a non-catalytic action of carbonic anhydrase II. J. Physiol. 590, 2333–2351 10.1113/jphysiol.2011.220152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Noor S. I., Dietz S., Heidtmann H., Boone C. D., McKenna R., Deitmer J. W., and Becker H. M. (2015) Analysis of the binding moiety mediating the interaction between monocarboxylate transporters and carbonic anhydrase II. J. Biol. Chem. 290, 4476–4486 10.1074/jbc.M114.624577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gallagher S. M., Castorino J. J., Wang D., and Philp N. J. (2007) Monocarboxylate transporter 4 regulates maturation and trafficking of CD147 to the plasma membrane in the metastatic breast cancer cell line MDA-MB-231. Cancer Res. 67, 4182–4189 10.1158/0008-5472.CAN-06-3184 [DOI] [PubMed] [Google Scholar]
- 65. Isom D. G., Castañeda C. A., Cannon B. R., and García-Moreno B. (2011) Large shifts in pKa values of lysine residues buried inside a protein. Proc. Natl. Acad. Sci. U.S.A. 108, 5260–5265 10.1073/pnas.1010750108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shinobu A., and Agmon N. (2009) Mapping proton wires in proteins: carbonic anhydrase and GFP chromophore biosynthesis. J. Phys. Chem. A. 113, 7253–7266 10.1021/jp8102047 [DOI] [PubMed] [Google Scholar]
- 67. Ames S., Pastorekova S., and Becker H. M. (2018) The proteoglycan-like domain of carbonic anhydrase IX mediates non-catalytic facilitation of lactate transport in cancer cells. Oncotarget 9, 27940–27957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bruce B. L. J., Ring S. M., Anstee D. J., Reid M. E., Wilkinson S., and Tanner M. J. A. (1995) Changes in the blood group wright antigens are associated with a mutation at amino acid 658 in human erythrocyte band 3: a site of interaction between band 3 and glycophorin A under certain conditions. Blood 85, 541–547 [PubMed] [Google Scholar]
- 69. Jarolim P., Rubin H. L., Zakova D., Storry J., and Reid M. E. (1998) Characterization of seven low incidence blood group antigens carried by erythrocyte band 3 protein. Blood 92, 4836–4843 [PubMed] [Google Scholar]
- 70. Wilson M. C., Kraus M., Marzban H., Sarna J. R., Wang Y., Hawkes R., Halestrap A. P., and Beesley P. W. (2013) The neuroplastin adhesion molecules are accessory proteins that chaperone the monocarboxylate transporter MCT2 to the neuronal cell surface. PLoS ONE 8, e78654 10.1371/journal.pone.0078654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schwede T., Kopp J., Guex N., and Peitsch M. C. (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 10.1093/nar/gkg520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 73. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 10.1107/S0907444998003254 [DOI] [PubMed] [Google Scholar]
- 74. Shnitsar V., Li J., Li X., Calmettes C., Basu A., Casey J. R., Moraes T. F., and Reithmeier R. A. (2013) A substrate access tunnel in the cytosolic domain is not an essential feature of the solute carrier 4 (SLC4) family of bicarbonate transporters. J. Biol. Chem. 288, 33848–33860 10.1074/jbc.M113.511865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sterling D., and Casey J. R. (1999) Transport activity of AE3 chloride/bicarbonate anion-exchange proteins and their regulation by intracellular pH. Biochem. J. 344, 221–229 10.1042/bj3440221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Becker H. M., Bröer S., and Deitmer J. W. (2004) Facilitated lactate transport by MCT1 when coexpressed with the sodium bicarbonate cotransporter (NBC) in Xenopus oocytes. Biophys. J. 86, 235–247 10.1016/S0006-3495(04)74099-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Becker H. M. (2014) Transport of lactate: characterization of the transporters involved in transport at the plasma membrane by heterologous protein expression in Xenopus oocytes. Neuromethods 90, 25–43 10.1007/978-1-4939-1059-5_2 [DOI] [Google Scholar]
- 78. Deitmer J. W. (1991) Electrogenic sodium-dependent bicarbonate secretion by glial cells of the leech central nervous system. J. Gen. Physiol. 98, 637–655 10.1085/jgp.98.3.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Stams T., Nair S. K., Okuyama T., Waheed A., Sly W. S., and Christianson D. W. (1996) Crystal structure of the secretory form of membrane-associated human carbonic anhydrase IV at 2.8-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 93, 13589–13594 10.1073/pnas.93.24.13589 [DOI] [PMC free article] [PubMed] [Google Scholar]